bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1102_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
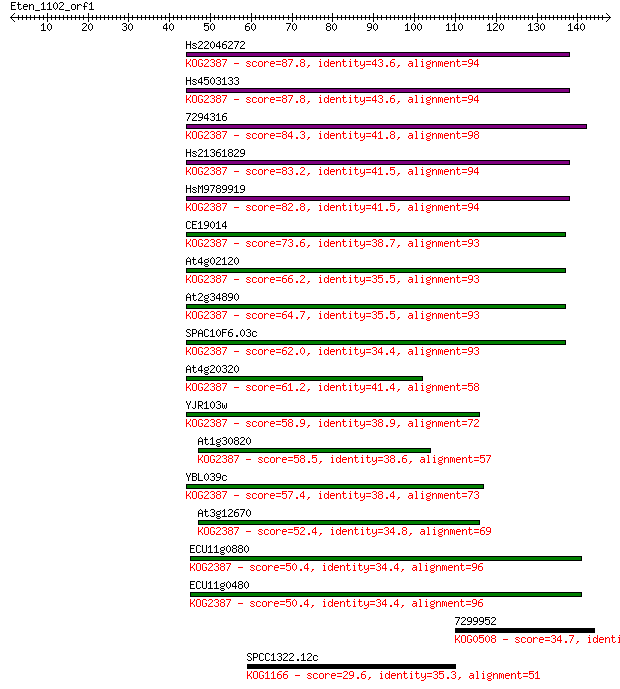
Hs22046272 87.8 6e-18
Hs4503133 87.8 7e-18
7294316 84.3 7e-17
Hs21361829 83.2 2e-16
HsM9789919 82.8 2e-16
CE19014 73.6 1e-13
At4g02120 66.2 2e-11
At2g34890 64.7 7e-11
SPAC10F6.03c 62.0 4e-10
At4g20320 61.2 6e-10
YJR103w 58.9 3e-09
At1g30820 58.5 5e-09
YBL039c 57.4 9e-09
At3g12670 52.4 3e-07
ECU11g0880 50.4 1e-06
ECU11g0480 50.4 1e-06
7299952 34.7 0.073
SPCC1322.12c 29.6 2.4
> Hs22046272
Length=591
Score = 87.8 bits (216), Expect = 6e-18, Method: Composition-based stats.
Identities = 41/94 (43%), Positives = 57/94 (60%), Gaps = 3/94 (3%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ELRG GLSPD V CR PL + ++KIS+F V+P+ V VH+ +IY+VPL+LE Q
Sbjct 201 VRELRGLGLSPDLVVCRCSNPLDTSVKEKISMFCHVEPEQVICVHDVSSIYRVPLLLEEQ 260
Query 104 GLSRRLIERLGLEASGRRRDAAASLEVWRRMAER 137
G+ + RL L R L W+ MA+R
Sbjct 261 GVVDYFLRRLDLPI---ERQPRKMLMKWKEMADR 291
> Hs4503133
Length=591
Score = 87.8 bits (216), Expect = 7e-18, Method: Composition-based stats.
Identities = 41/94 (43%), Positives = 57/94 (60%), Gaps = 3/94 (3%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ELRG GLSPD V CR PL + ++KIS+F V+P+ V VH+ +IY+VPL+LE Q
Sbjct 201 VRELRGLGLSPDLVVCRCSNPLDTSVKEKISMFCHVEPEQVICVHDVSSIYRVPLLLEEQ 260
Query 104 GLSRRLIERLGLEASGRRRDAAASLEVWRRMAER 137
G+ + RL L R L W+ MA+R
Sbjct 261 GVVDYFLRRLDLPI---ERQPRKMLMKWKEMADR 291
> 7294316
Length=627
Score = 84.3 bits (207), Expect = 7e-17, Method: Composition-based stats.
Identities = 41/98 (41%), Positives = 57/98 (58%), Gaps = 2/98 (2%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ELRG GLSPD + CRSE+P+ ++KIS F V P V +H+ +IY VPL++E
Sbjct 200 VRELRGCGLSPDLIVCRSEKPIGLEVKEKISNFCHVGPDQVICIHDLNSIYHVPLLMEQN 259
Query 104 GLSRRLIERLGLEASGRRRDAAASLEVWRRMAERLRIV 141
G+ L ERL L +R L+ WR +A R V
Sbjct 260 GVIEYLNERLQLNIDMSKR--TKCLQQWRDLARRTETV 295
> Hs21361829
Length=586
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 39/95 (41%), Positives = 59/95 (62%), Gaps = 5/95 (5%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ LRG GLSPD + CRS P+ A ++KIS+F V P+ V +H+ + Y+VP++LE Q
Sbjct 201 VRALRGLGLSPDLIVCRSSSPIEMAVKEKISMFCHVNPEQVICIHDVSSTYRVPVLLEEQ 260
Query 104 GLSRRLIERLGLEASGRRRDAAASLEV-WRRMAER 137
+ + ERL L D+A++L WR MA+R
Sbjct 261 SIVKYFKERLHLPIG----DSASNLLFKWRNMADR 291
> HsM9789919
Length=586
Score = 82.8 bits (203), Expect = 2e-16, Method: Composition-based stats.
Identities = 39/95 (41%), Positives = 59/95 (62%), Gaps = 5/95 (5%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ LRG GLSPD + CRS P+ A ++KIS+F V P+ V +H+ + Y+VP++LE Q
Sbjct 201 VRALRGLGLSPDLIVCRSSTPIEMAVKEKISMFCHVNPEQVICIHDVSSTYRVPVLLEEQ 260
Query 104 GLSRRLIERLGLEASGRRRDAAASLEV-WRRMAER 137
+ + ERL L D+A++L WR MA+R
Sbjct 261 SIVKYFKERLHLPIG----DSASNLLFKWRNMADR 291
> CE19014
Length=599
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 36/95 (37%), Positives = 55/95 (57%), Gaps = 2/95 (2%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ LR AGL PD + CRS E L R+KI+ F + + V VH+ NIYKVPL+L+ Q
Sbjct 207 VRHLRAAGLVPDLLICRSTEKLDPHLREKIAGFGMMDLEQVVGVHDVSNIYKVPLLLQQQ 266
Query 104 GLSRRLIERLGLE--ASGRRRDAAASLEVWRRMAE 136
G+ + +RL L + R+D ++ W ++E
Sbjct 267 GVLEAITQRLRLSEISENIRKDLKFNMNHWTNLSE 301
> At4g02120
Length=539
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 33/93 (35%), Positives = 50/93 (53%), Gaps = 8/93 (8%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ELR GL+P F+ CRS +PL E+ + K+S F V N+ ++H+ NI+ VPL+L
Sbjct 201 VRELRALGLTPHFLACRSAQPLLESTKAKLSQFCHVAAANILNIHDVPNIWHVPLLLRCN 260
Query 104 GLSRRLIERLGLEASGRRRDAAASLEVWRRMAE 136
+LI A L+ W +MAE
Sbjct 261 TKFNKLIS--------FSVATAPDLDSWNKMAE 285
> At2g34890
Length=597
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 33/93 (35%), Positives = 52/93 (55%), Gaps = 4/93 (4%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
+++LRG GL+P+ + CRS + L E + K+S F V QN+FS+ + NI+ +PL+L+ Q
Sbjct 201 IKDLRGFGLTPNIIACRSTKALEENVKAKLSRFCYVPIQNIFSLCDVPNIWHIPLLLKEQ 260
Query 104 GLSRRLIERLGLEASGRRRDAAASLEVWRRMAE 136
+ + L L + SLE W M E
Sbjct 261 KAHEAISKVLNLSGIAKE----PSLEKWASMVE 289
> SPAC10F6.03c
Length=600
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 32/97 (32%), Positives = 54/97 (55%), Gaps = 5/97 (5%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
+++LR G++PD + CR ++PL ++ KISLF V P+ V +VH+ + Y VP +LE +
Sbjct 199 IRDLRSLGITPDLIACRCKQPLEKSVIDKISLFCHVGPEQVLAVHDVSSTYHVPQLLEDK 258
Query 104 GLSRRLIERLGLEASGRRRDAAASLE----VWRRMAE 136
L I R L+ R+ A + E W+ + +
Sbjct 259 LLEYLKI-RFALDKISVSRELALAGENMWSSWKHLTQ 294
> At4g20320
Length=553
Score = 61.2 bits (147), Expect = 6e-10, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 42/58 (72%), Gaps = 0/58 (0%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILE 101
+++LRG GLSP+ + CRS +PL + + K+S F V +NV ++++C NI+ +PL+L+
Sbjct 197 VRDLRGLGLSPNILACRSTKPLEDNVKAKLSQFCHVPMENVVTLYDCPNIWHIPLLLK 254
> YJR103w
Length=564
Score = 58.9 bits (141), Expect = 3e-09, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
+++LR GL PD + CR E L+ + KI++F V P+ V +VH+ + Y VPL+L Q
Sbjct 199 IKDLRSLGLIPDMIACRCSEELNRSTIDKIAMFCHVGPEQVVNVHDVNSTYHVPLLLLKQ 258
Query 104 GLSRRLIERLGL 115
+ L RL L
Sbjct 259 HMIDYLHSRLKL 270
> At1g30820
Length=600
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 41/57 (71%), Gaps = 0/57 (0%)
Query 47 LRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
LRG GL+PD + CRS +PL + ++K++ F V + +F++++ NI+++PL+L+ Q
Sbjct 204 LRGLGLTPDILACRSTKPLEDNVKEKLAQFCHVPLEYIFTLYDVPNIWRIPLLLKDQ 260
> YBL039c
Length=579
Score = 57.4 bits (137), Expect = 9e-09, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 41/73 (56%), Gaps = 0/73 (0%)
Query 44 LQELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQ 103
++ LR GL PD + CR E L + KI++F V P+ V +VH+ + Y VPL+L Q
Sbjct 199 IKGLRSLGLVPDMIACRCSETLDKPTIDKIAMFCHVGPEQVVNVHDVNSTYHVPLLLLEQ 258
Query 104 GLSRRLIERLGLE 116
+ L RL L+
Sbjct 259 KMIDYLHARLKLD 271
> At3g12670
Length=591
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 24/69 (34%), Positives = 39/69 (56%), Gaps = 0/69 (0%)
Query 47 LRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQGLS 106
LR GL+P+ + CRS + L E + K+S F V N+ ++++ NI+ VPL+L Q
Sbjct 204 LRSLGLTPNILACRSTKALEENVKTKLSQFCHVPEVNIVTLYDVPNIWHVPLLLRDQKAH 263
Query 107 RRLIERLGL 115
++ L L
Sbjct 264 EAILRELNL 272
> ECU11g0880
Length=535
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 33/97 (34%), Positives = 47/97 (48%), Gaps = 1/97 (1%)
Query 45 QELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQG 104
+ R GL+ D V CR + R+KIS V+ +NV + N E++Y P+ LE G
Sbjct 205 RNFRRFGLNYDIVICRGRREPNMETRRKISTSCWVKEENVLGLPNLESVYLAPMFLEKHG 264
Query 105 LSRRLIERLGLEASGR-RRDAAASLEVWRRMAERLRI 140
+ L LGL+ G RR V RR + +RI
Sbjct 265 IVEALNRILGLDDKGMDRRMLDIFSMVGRRHRDGVRI 301
> ECU11g0480
Length=535
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 33/97 (34%), Positives = 47/97 (48%), Gaps = 1/97 (1%)
Query 45 QELRGAGLSPDFVFCRSEEPLSEAARQKISLFAQVQPQNVFSVHNCENIYKVPLILEAQG 104
+ R GL+ D V CR + R+KIS V+ +NV + N E++Y P+ LE G
Sbjct 205 RNFRRFGLNYDIVICRGRREPNMETRRKISTSCWVKEENVLGLPNLESVYLAPMFLEKHG 264
Query 105 LSRRLIERLGLEASGR-RRDAAASLEVWRRMAERLRI 140
+ L LGL+ G RR V RR + +RI
Sbjct 265 IVEALNRILGLDDKGMDRRMLDIFSMVGRRHRDGVRI 301
> 7299952
Length=662
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 110 IERLGLEASGRRRDAAASLEVWRRMAERLRIVPP 143
+E LG R+RD AA+L +WRR E + PP
Sbjct 246 LELLGATYVDRKRDMAAALNLWRRALEERAVEPP 279
> SPCC1322.12c
Length=1044
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 6/57 (10%)
Query 59 CRSEEPLSEAARQKISLFAQVQPQNVFSVH------NCENIYKVPLILEAQGLSRRL 109
CR++ LS + F +V P+N SVH N IYK P+ ++ +R L
Sbjct 289 CRAQRYLSSIQPNTAASFPKVVPKNEISVHHDSSSSNVSPIYKNPVAEQSDTPTRSL 345
Lambda K H
0.330 0.144 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40