bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1127_orf2
Length=250
Score E
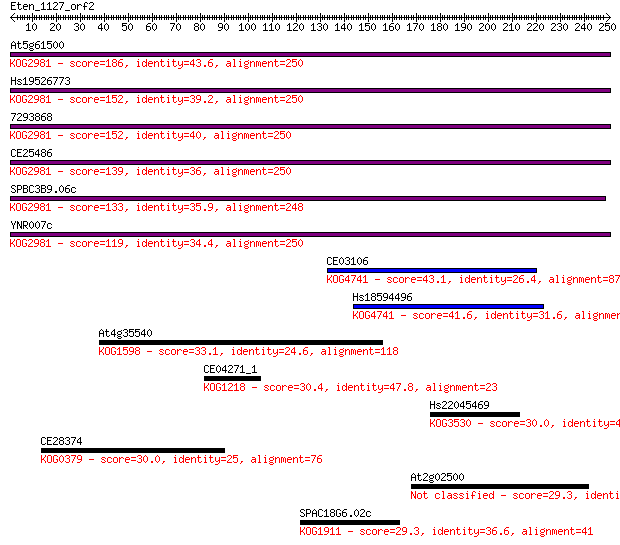
Sequences producing significant alignments: (Bits) Value
At5g61500 186 3e-47
Hs19526773 152 6e-37
7293868 152 7e-37
CE25486 139 7e-33
SPBC3B9.06c 133 3e-31
YNR007c 119 5e-27
CE03106 43.1 6e-04
Hs18594496 41.6 0.001
At4g35540 33.1 0.62
CE04271_1 30.4 3.4
Hs22045469 30.0 4.3
CE28374 30.0 4.5
At2g02500 29.3 7.4
SPAC18G6.02c 29.3 7.6
> At5g61500
Length=311
Score = 186 bits (473), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 109/261 (41%), Positives = 147/261 (56%), Gaps = 18/261 (6%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA------R 54
D LV K PTW WE + +LP DKQFL+TR V C RR + A
Sbjct 43 DNLVSKCPTWSWESGDASKRKPYLPSDKQFLITRNVPCLRRAASVAEDYEAAGGEVLVDD 102
Query 55 EDAEGWVL----PVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNL 110
ED +GW+ P EE ++ E N ++ P G D +PD++
Sbjct 103 EDNDGWLATHGKPKGKEEDNLPSMDALDINEKNTIQSIPTYFGGEEDDD----IPDMEEF 158
Query 111 RDLDSLLCEDDPACADSSTSYFVRNAPDAD-VLSVRTYDLSITYDKYFQTPRLWLFGYNE 169
+ D+++ E+DPA S+ Y V + PD D +L RTYDLSITYDKY+QTPR+WL GY+E
Sbjct 159 DEADNVV-ENDPATLQST--YLVAHEPDDDNILRTRTYDLSITYDKYYQTPRVWLTGYDE 215
Query 170 SGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPTASIHPCKHAQVMKKVVDHWRSQGVE 229
S L P + ED+ D+A KTVT++ HP AS+HPC+H VMKK++D S+GVE
Sbjct 216 SRMLLQPELVMEDVSQDHARKTVTIEDHPHLPGKHASVHPCRHGAVMKKIIDVLMSRGVE 275
Query 230 PRPDLALIVLLKFISSVVPTI 250
P D L + LKF++SV+PTI
Sbjct 276 PEVDKYLFLFLKFMASVIPTI 296
> Hs19526773
Length=314
Score = 152 bits (384), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 98/270 (36%), Positives = 142/270 (52%), Gaps = 29/270 (10%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA---REDA 57
D LV PTWQW ++ ++LP KQFLVT+ V C++R + +E S EA +D
Sbjct 44 DHLVHHCPTWQWATGEELKVKAYLPTGKQFLVTKNVPCYKRCKQMEYSDELEAIIEEDDG 103
Query 58 EG-WVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDL--- 113
+G WV + + G T +++ + ++R AL + + D
Sbjct 104 DGGWV---------DTYHNTGITGITEAVKEITLENKDNIRLQDCSALCEEEEDEDEGEA 154
Query 114 -------DSLLCEDDPACADS-STSYFVRNAPDAD----VLSVRTYDLSITYDKYFQTPR 161
+S L E D A D+ + DA +L RTYDL ITYDKY+QTPR
Sbjct 155 ADMEEYEESGLLETDEATLDTRKIVEACKAKTDAGGEDAILQTRTYDLYITYDKYYQTPR 214
Query 162 LWLFGYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCT-GVPTASIHPCKHAQVMKKVV 220
LWLFGY+E PLT ++EDI D+ KTVT++ HP P S+HPC+HA+VMKK++
Sbjct 215 LWLFGYDEQRQPLTVEHMYEDISQDHVKKTVTIENHPHLPPPPMCSVHPCRHAEVMKKII 274
Query 221 DHWRSQGVEPRPDLALIVLLKFISSVVPTI 250
+ G E + L++ LKF+ +V+PTI
Sbjct 275 ETVAEGGGELGVHMYLLIFLKFVQAVIPTI 304
> 7293868
Length=330
Score = 152 bits (384), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 100/278 (35%), Positives = 142/278 (51%), Gaps = 30/278 (10%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIE----ASLAAEARED 56
D LV PTWQW + +LP DKQFL+TR V C+RR + +E +L E D
Sbjct 44 DHLVHHCPTWQWAAGDETKTKPYLPKDKQFLITRNVPCYRRCKQMEYVGEETLVEEESGD 103
Query 57 AEGWV----------LPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPD 106
GWV +E + E + + T +K G+G D D
Sbjct 104 G-GWVETHQLNDDGTTQLEDKICELTMEETKEEMHTPDSDKSAPGAGGQAEDEDDDEAID 162
Query 107 LKNLRDLDSLLCEDDPACA--------DSSTSYFVRNAPDAD-----VLSVRTYDLSITY 153
+ + + +L DPA A ++ S + DA+ VL RTYDL I+Y
Sbjct 163 MDDFEE-SGMLELVDPAVATTTRKPEPEAKASPVAAASGDAEASGDSVLHTRTYDLHISY 221
Query 154 DKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAKTVTVDPHP-CTGVPTASIHPCKH 212
DKY+QTPRLW+ GY+E PLT +++ED+ D+A KTVT++ HP G AS+HPC+H
Sbjct 222 DKYYQTPRLWVVGYDEQRKPLTVEQMYEDVSQDHAKKTVTMESHPHLPGPNMASVHPCRH 281
Query 213 AQVMKKVVDHWRSQGVEPRPDLALIVLLKFISSVVPTI 250
A +MKK++ G + L LI+ LKF+ +V+PTI
Sbjct 282 ADIMKKIIQTVEEGGGQLGVHLYLIIFLKFVQTVIPTI 319
> CE25486
Length=305
Score = 139 bits (349), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 90/265 (33%), Positives = 129/265 (48%), Gaps = 28/265 (10%)
Query 1 DLLVFKFPTWQWEPAVG-KRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEA---RED 56
D LV PTW+W A +I ++LP DKQFL+TR V CH+R + +E E ED
Sbjct 44 DHLVHHCPTWKWAGASDPSKIRTFLPIDKQFLITRNVPCHKRCKQMEYDEKLEKIINEED 103
Query 57 AE-------GWVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKN 109
E GWV + + + PP S D D
Sbjct 104 GEYQTSDETGWV-----DTHHYEKEHEKNEEKEQSTAPPPAAPEDSDDDDEEPLDLDELG 158
Query 110 LRDL----DSLLCEDDPACADSSTSYFVRNAPDADVLSVRTYDLSITYDKYFQTPRLWLF 165
L D + + E P A + S +V VRTYDL I YDKY+Q PRL+L
Sbjct 159 LDDDEEDPNRFVVEKKPLAAGNDNS--------GEVEKVRTYDLHICYDKYYQVPRLFLM 210
Query 166 GYNESGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPTASIHPCKHAQVMKKVVDHWRS 225
GY+E+ PLT + +ED D++ KT+TV+ HP + ++HPCKHA++MK++++ +
Sbjct 211 GYDENRRPLTVEQTYEDFSADHSNKTITVEAHPSVDLTMPTVHPCKHAEMMKRLINQYAE 270
Query 226 QGVEPRPDLALIVLLKFISSVVPTI 250
G L + LKF+ +V+PTI
Sbjct 271 SGKVLGVHEYLFLFLKFVQAVIPTI 295
> SPBC3B9.06c
Length=275
Score = 133 bits (335), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 89/256 (34%), Positives = 131/256 (51%), Gaps = 39/256 (15%)
Query 1 DLLVFKFPTWQWEPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEAREDAEGW 60
D LV KFPTW WE G RI +LP DKQ+LVTR V C +R +I + E W
Sbjct 41 DYLVSKFPTWSWE--CGDRIRGFLPKDKQYLVTRHVFCVQRNINIGVN---------EEW 89
Query 61 VLPVEAEEPEE------VLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDLD 114
V +E ++ ++S +T+ + G S + G LP LK+ D D
Sbjct 90 V-DIETDDTRNKDDDQDDDAISSIHSDTSDIASAERLKGQSKELSDSGPLP-LKDEEDDD 147
Query 115 SLLCEDDPACADSSTSYFVRNAPDADVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPL 174
++ P + Y YDL I YDKY++TPRL+L G+N G L
Sbjct 148 QMVS---PVIKEDEGRY---------------YDLYIVYDKYYRTPRLFLRGWNAGGQLL 189
Query 175 TPVEIFEDILTDYAAKTVTVD--PHPCTGVPTASIHPCKHAQVMKKVVDHWRSQGVEPRP 232
T +I+ED+ ++A KTVT++ PH + AS+HPCKHA V+ K++ R + R
Sbjct 190 TMKDIYEDVSGEHAGKTVTMEPFPHYHSHNTMASVHPCKHASVLLKLIKQHRERNDPIRV 249
Query 233 DLALIVLLKFISSVVP 248
D +++ LKF+S+++P
Sbjct 250 DQYMVLFLKFVSTMLP 265
> YNR007c
Length=310
Score = 119 bits (299), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 86/283 (30%), Positives = 128/283 (45%), Gaps = 52/283 (18%)
Query 1 DLLVFKFPTWQW-EPAVGKRINSWLPPDKQFLVTRGVACHRRVRDIEASLAAEAREDAEG 59
D L FPTW+W E + +LP +KQFL+ R V C +R
Sbjct 38 DYLCHMFPTWKWNEESSDISYRDFLPKNKQFLIIRKVPCDKRAEQC-------------- 83
Query 60 WVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLRDTSGGALPDLKNLRDLDSLLCE 119
VE E P+ ++ G+ + + + G ++ T G D ++ D+D L+ +
Sbjct 84 ----VEVEGPDVIMKGFAEDGDEDDVLEYIGSETEHVQSTPAGGTKD-SSIDDIDELIQD 138
Query 120 DDPACADSSTSYFVRNAPDA---DVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTP 176
+ D + NA D+ R YDL I Y ++ P++++ G+N +G+PL+P
Sbjct 139 MEIKEEDENDDTEEFNAKGGLAKDMAQERYYDLYIAYSTSYRVPKMYIVGFNSNGSPLSP 198
Query 177 VEIFEDILTDYAAKTVTVDPHPC--TGVPTASIHPCKHAQVMKKVVDHWRS--------- 225
++FEDI DY KT T++ P V + SIHPCKHA VMK ++D R
Sbjct 199 EQMFEDISADYRTKTATIEKLPFYKNSVLSVSIHPCKHANVMKILLDKVRVVRQRRRKEL 258
Query 226 ------QGV------------EPRPDLALIVLLKFISSVVPTI 250
GV R D LIV LKFI+SV P+I
Sbjct 259 QEEQELDGVGDWEDLQDDIDDSLRVDQYLIVFLKFITSVTPSI 301
> CE03106
Length=173
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 44/95 (46%), Gaps = 11/95 (11%)
Query 133 VRNAPDADVLSVRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAK-- 190
++ D V++ T+ I Y+ +Q P +W + +G+PL + D+L +
Sbjct 55 LKTLSDGRVITSETH---ILYNSTYQVPTIWFNFFENNGSPLPFRTVIRDVLNISETEES 111
Query 191 ------TVTVDPHPCTGVPTASIHPCKHAQVMKKV 219
++ HP GV +IHPC + +MK++
Sbjct 112 EASIRSRISHYEHPFMGVLYYNIHPCNTSNIMKEL 146
> Hs18594496
Length=220
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 38/86 (44%), Gaps = 7/86 (8%)
Query 144 VRTYDLSITYDKYFQTPRLWLFGYNESGTPLTPVEIFEDILTDYAAK-------TVTVDP 196
V Y+ + Y +Q P L+ G PLT +I+E + Y + T+T
Sbjct 93 VIKYEYHVLYSCSYQVPVLYFRASFLDGRPLTLKDIWEGVHECYKMRLLQGPWDTITQQE 152
Query 197 HPCTGVPTASIHPCKHAQVMKKVVDH 222
HP G P +HPCK + M V+ +
Sbjct 153 HPILGQPFFVLHPCKTNEFMTPVLKN 178
> At4g35540
Length=527
Score = 33.1 bits (74), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 50/119 (42%), Gaps = 3/119 (2%)
Query 38 CHRRVRDIEASLAAEAREDAEGWVLPVEAEEPEEVLSVSVSGGETNPLEKPPGGSGSSLR 97
C R +++ L A E W V + + + E ++K G+G L
Sbjct 246 CKTRYKELSEKLVKVAEEVGLPWAKDVTVKNVLKHSGTLFALMEAKSMKKRKQGTGKELV 305
Query 98 DTSGGALPDL-KNLRDLDSLLCEDDPACADSSTSYFVRNAPDADVLSVRTYDLSITYDK 155
T G + DL + +S+ C DD A D+ + YF + LS+ YD +I+ ++
Sbjct 306 RTDGFCVEDLVMDCLSKESMYCYDDDARQDTMSRYF--DVEGERQLSLCNYDDNISENQ 362
> CE04271_1
Length=2510
Score = 30.4 bits (67), Expect = 3.4, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 82 TNPLEKPPGGSGSSLRDTSGGAL 104
+ P+ PPGGS S+LR+ +GG++
Sbjct 1694 SEPVTSPPGGSCSNLRECTGGSV 1716
> Hs22045469
Length=633
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 19/42 (45%), Gaps = 5/42 (11%)
Query 176 PVEIFEDILTDYAAKTVTVDPHPCTGVPTASIH-----PCKH 212
PV F +L + +T VDPHPC + H CKH
Sbjct 415 PVAEFNLLLKAHTLETYGVDPHPCKKKAMLAFHTSTPAACKH 456
> CE28374
Length=580
Score = 30.0 bits (66), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 19/78 (24%), Positives = 38/78 (48%), Gaps = 2/78 (2%)
Query 14 PAVGKRINSWLPPDKQFLV--TRGVACHRRVRDIEASLAAEAREDAEGWVLPVEAEEPEE 71
P G+ I + P K +++ T GV + V + + EA+ ++E + E E
Sbjct 155 PLYGQAICAGEIPGKYYIIGGTEGVRFNFDVHSLTMKVNPEAKHESEKFTWHSELVSQNE 214
Query 72 VLSVSVSGGETNPLEKPP 89
++++++ ETN L+ P
Sbjct 215 IVTLNIRTRETNELQTIP 232
> At2g02500
Length=278
Score = 29.3 bits (64), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 38/75 (50%), Gaps = 6/75 (8%)
Query 168 NESGTPLTPVEIFEDILTDYAAKTVTVDPHPCTGVPT-ASIHPCKHAQVMKKVVDHWRSQ 226
++S PL E E +L D +A V GVP A+I +++KK + +S+
Sbjct 177 HDSARPLVNTEDVEKVLKDGSAVGAAV-----LGVPAKATIKEVIKPELLKKGFELVKSE 231
Query 227 GVEPRPDLALIVLLK 241
G+E D++++ LK
Sbjct 232 GLEVTDDVSIVEYLK 246
> SPAC18G6.02c
Length=960
Score = 29.3 bits (64), Expect = 7.6, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Query 122 PACADSSTSYFVRNAPDADVLSVRTYDLSITYDKYFQTPRL 162
P+ D SYFV N PD +L ++ +L T +Y P L
Sbjct 848 PSIVDMVKSYFVTNNPDKSLLEIQ--NLLNTLQRYLTNPAL 886
Lambda K H
0.317 0.135 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5116459104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40