bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
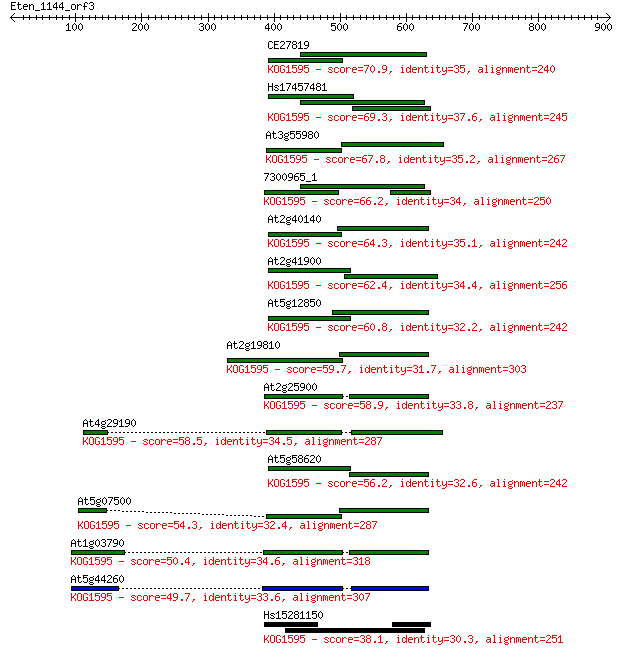
Query= Eten_1144_orf3
Length=908
Score E
Sequences producing significant alignments: (Bits) Value
CE27819 70.9 1e-11
Hs17457481 69.3 3e-11
At3g55980 67.8 1e-10
7300965_1 66.2 4e-10
At2g40140 64.3 1e-09
At2g41900 62.4 4e-09
At5g12850 60.8 1e-08
At2g19810 59.7 3e-08
At2g25900 58.9 5e-08
At4g29190 58.5 7e-08
At5g58620 56.2 3e-07
At5g07500 54.3 1e-06
At1g03790 50.4 2e-05
At5g44260 49.7 3e-05
Hs15281150 38.1 0.093
> CE27819
Length=704
Score = 70.9 bits (172), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 65/219 (29%), Positives = 86/219 (39%), Gaps = 35/219 (15%)
Query 440 DSSSLQCSKGDACDKSHNRH---ELLYHPSIFKQRFCSSYATRNGTERCGR-GHFCAFAH 495
D ++ C G+ C H E YH +K C G +C + G CAFAH
Sbjct 85 DENTGICPDGEDCIFLHRVSGDVERKYHLRYYKTAQCVHPTDARG--QCVKNGAHCAFAH 142
Query 496 TREEVRAPLF---------------------TVQEETCPSSDFFMQHFKTVWC--PYGVQ 532
T ++R P+F +++ S D + +KT C P +
Sbjct 143 TANDIRPPMFDQHEVGFSTVVDGEGRDKTSFVIEDPQWHSQDHVLSCYKTEQCRKPARLC 202
Query 533 HDWYTCVYAHTYQDCRRSPQV-GYGSEPCPAWNKDVHSADYSRRCPHGTLCPYSHGSKEQ 591
Y C + H +D RR P + Y S PCPA K + C G C Y H EQ
Sbjct 203 RQGYACPFYHNSKDRRRPPALYKYRSTPCPA-AKTIDEWLDPDICEAGDNCQYCHTRTEQ 261
Query 592 LYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASE 630
+HP YK+ C D CPR V CAF H SE
Sbjct 262 QFHPEIYKSTKCNDMLEH----GYCPRAVFCAFAHHDSE 296
Score = 64.7 bits (156), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 53/120 (44%), Gaps = 15/120 (12%)
Query 391 LSSFKVFPCRHKHSVSHDKKYCPFYHNFRDKRRFPVTYR--------AEQCEEHFDLDSS 442
LS +K CR + CPFYHN +D+RR P Y+ A+ +E D D
Sbjct 187 LSCYKTEQCRKPARLCRQGYACPFYHNSKDRRRPPALYKYRSTPCPAAKTIDEWLDPDI- 245
Query 443 SLQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVRA 502
C GD C H R E +HP I+K C+ C R FCAFAH E+ A
Sbjct 246 ---CEAGDNCQYCHTRTEQQFHPEIYKSTKCNDMLEHG---YCPRAVFCAFAHHDSELHA 299
> Hs17457481
Length=412
Score = 69.3 bits (168), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 46/137 (33%), Positives = 65/137 (47%), Gaps = 11/137 (8%)
Query 391 LSSFKVFPCRHKHSVSHDKKYCPFYHNFRDKRRFPV--TYRAEQCE--EHFDLDSSSLQC 446
L ++K PC+ + CP+YHN +D+RR P YR+ C +H D +C
Sbjct 213 LGNYKTEPCKKPPRLCRQGYACPYYHNSKDRRRSPRKHKYRSSPCPNVKHGDEWGDPGKC 272
Query 447 SKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVRA---- 502
GDAC H R E +HP I+K C+ ++G+ C RG FCAFAH + +
Sbjct 273 ENGDACQYCHTRTEQQFHPEIYKSTKCND-MQQSGS--CPRGPFCAFAHVEQPPLSDDLQ 329
Query 503 PLFTVQEETCPSSDFFM 519
P V T P +M
Sbjct 330 PSSAVSSPTQPGPVLYM 346
Score = 64.7 bits (156), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 67/232 (28%), Positives = 93/232 (40%), Gaps = 52/232 (22%)
Query 440 DSSSLQCSKGDACDKSHNRH---ELLYHPSIFKQRFCSSYATRNGTERCGR-GHFCAFAH 495
D ++ C +GD C H E YH +K C G C + G CAFAH
Sbjct 94 DEATGLCPEGDECPFLHRTTGDTERRYHLRYYKTGICIHETDSKGN--CTKNGLHCAFAH 151
Query 496 TREEVRAPLFTVQE-------------------------------ETCPSSD-------F 517
++R+P++ ++E E S + +
Sbjct 152 GPHDLRSPVYDIRELQAMEALQNGQTTVEGSIEGQSAGAASHAMIEKILSEEPRWQETAY 211
Query 518 FMQHFKTVWC--PYGVQHDWYTCVYAHTYQDCRRSPQV-GYGSEPCPAWNKDVHSADYSR 574
+ ++KT C P + Y C Y H +D RRSP+ Y S PCP D
Sbjct 212 VLGNYKTEPCKKPPRLCRQGYACPYYHNSKDRRRSPRKHKYRSSPCPNVKHGDEWGD-PG 270
Query 575 RCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYH 626
+C +G C Y H EQ +HP YK+ C D + +SG SCPRG CAF H
Sbjct 271 KCENGDACQYCHTRTEQQFHPEIYKSTKCNDMQ---QSG-SCPRGPFCAFAH 318
Score = 49.7 bits (117), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 45/130 (34%), Positives = 60/130 (46%), Gaps = 17/130 (13%)
Query 518 FMQHFKTVWCPYGVQHDW-----YTCVYAH-TYQDCRRSPQVGYGSEPCPAWNKDVHSAD 571
+++ F+T CP VQH YTC + H Q RRS + G+ ++ DV+
Sbjct 36 YLKEFRTEQCPLFVQHKCTQHRPYTCFHWHFVNQRRRRSIRRRDGTF---NYSPDVYCTK 92
Query 572 YSRR---CPHGTLCPYSH---GSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFY 625
Y CP G CP+ H G E+ YH YYKT C+ D GN G+ CAF
Sbjct 93 YDEATGLCPEGDECPFLHRTTGDTERRYHLRYYKTGICI--HETDSKGNCTKNGLHCAFA 150
Query 626 HEASERRWPV 635
H + R PV
Sbjct 151 HGPHDLRSPV 160
> At3g55980
Length=586
Score = 67.8 bits (164), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 51/156 (32%), Positives = 65/156 (41%), Gaps = 19/156 (12%)
Query 502 APLFTVQEETCPSSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYGSEP 559
A L + E S +F M FK C HDW C + H ++ RR + Y P
Sbjct 203 ASLPDINEGVYGSDEFRMYSFKVKPCSRAYSHDWTECAFVHPGENARRRDPRKYPYTCVP 262
Query 560 CPAWNKDVHSADYSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRG 619
CP + K CP G C Y+HG E HP+ YKT C KD +G C R
Sbjct 263 CPEFRKG--------SCPKGDSCEYAHGVFESWLHPAQYKTRLC-----KDETG--CARK 307
Query 620 VLCAFYHEASERRWPVRMAIDYSSPLPPQRSAALQP 655
V C F H+ E R PV + + P S + P
Sbjct 308 V-CFFAHKREEMR-PVNASTGSAVAQSPFSSLEMMP 341
Score = 58.5 bits (140), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 43/118 (36%), Positives = 53/118 (44%), Gaps = 21/118 (17%)
Query 389 LNLSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDLDSSS 443
+ SFKV PC +S HD C F H RD R++P Y C E F S
Sbjct 218 FRMYSFKVKPCSRAYS--HDWTECAFVHPGENARRRDPRKYP--YTCVPCPE-FRKGS-- 270
Query 444 LQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVR 501
C KGD+C+ +H E HP+ +K R C C R C FAH REE+R
Sbjct 271 --CPKGDSCEYAHGVFESWLHPAQYKTRLCKDETG------CAR-KVCFFAHKREEMR 319
> 7300965_1
Length=541
Score = 66.2 bits (160), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 63/231 (27%), Positives = 89/231 (38%), Gaps = 58/231 (25%)
Query 440 DSSSLQCSKGDACDKSHNRH---ELLYHPSIFKQRFCS------SYATRNGTERCGRGHF 490
D ++ C +GD C H E YH +K C Y +NG
Sbjct 96 DETTGICPEGDECPYLHRTAGDTERRYHLRYYKTCMCVHDTDSRGYCVKNGLH------- 148
Query 491 CAFAHTREEVRAPLFTVQE-ETCPS-----------------------------SDFFMQ 520
CAFAH ++ R P++ ++E ET + +++ +
Sbjct 149 CAFAHGMQDQRPPVYDIKELETLQNAESTLDSTNALNALDKERNLMNEDPKWQDTNYVLA 208
Query 521 HFKTVWC--PYGVQHDWYTCVYAHTYQDCRRSP-QVGYGSEPCPAWNKDVHSADYSR--R 575
++KT C P + Y C H +D RRSP + Y S PCP H ++
Sbjct 209 NYKTEPCKRPPRLCRQGYACPQYHNSKDKRRSPRKYKYRSTPCPNVK---HGEEWGEPGN 265
Query 576 CPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYH 626
C G C Y H EQ +HP YK+ C D + CPR V CAF H
Sbjct 266 CEAGDNCQYCHTRTEQQFHPEIYKSTKCNDV----QQAGYCPRSVFCAFAH 312
Score = 58.9 bits (141), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 52/115 (45%), Gaps = 7/115 (6%)
Query 386 DGSLNLSSFKVFPCRHKHSVSHDKKYCPFYHNFRDKRRFP--VTYRAEQC--EEHFDLDS 441
D + L+++K PC+ + CP YHN +DKRR P YR+ C +H +
Sbjct 202 DTNYVLANYKTEPCKRPPRLCRQGYACPQYHNSKDKRRSPRKYKYRSTPCPNVKHGEEWG 261
Query 442 SSLQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHT 496
C GD C H R E +HP I+K C+ C R FCAFAH
Sbjct 262 EPGNCEAGDNCQYCHTRTEQQFHPEIYKSTKCNDVQQ---AGYCPRSVFCAFAHV 313
Score = 47.8 bits (112), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 31/63 (49%), Gaps = 5/63 (7%)
Query 576 CPHGTLCPYSH---GSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASERR 632
CP G CPY H G E+ YH YYKT CM D G G+ CAF H ++R
Sbjct 102 CPEGDECPYLHRTAGDTERRYHLRYYKT--CMCVHDTDSRGYCVKNGLHCAFAHGMQDQR 159
Query 633 WPV 635
PV
Sbjct 160 PPV 162
> At2g40140
Length=597
Score = 64.3 bits (155), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 46/139 (33%), Positives = 58/139 (41%), Gaps = 18/139 (12%)
Query 496 TREEVRAPLFTVQEETCPSSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QV 553
T+ A L + E + DF M FK C HDW C + H ++ RR +
Sbjct 194 TKYPADASLPDINEGVYGTDDFRMFSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKY 253
Query 554 GYGSEPCPAWNKDVHSADYSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSG 613
Y PCP + K CP G C Y+HG E HP+ Y+T C KD +G
Sbjct 254 PYTCVPCPEFRKG--------SCPKGDSCEYAHGVFESWLHPAQYRTRLC-----KDETG 300
Query 614 NSCPRGVLCAFYHEASERR 632
C R V C F H E R
Sbjct 301 --CARRV-CFFAHRRDELR 316
Score = 58.5 bits (140), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 54/116 (46%), Gaps = 21/116 (18%)
Query 391 LSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDLDSSSLQ 445
+ SFKV PC +S HD CPF H RD R++P Y C E F S
Sbjct 217 MFSFKVKPCSRAYS--HDWTECPFVHPGENARRRDPRKYP--YTCVPCPE-FRKGS---- 267
Query 446 CSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVR 501
C KGD+C+ +H E HP+ ++ R C C R C FAH R+E+R
Sbjct 268 CPKGDSCEYAHGVFESWLHPAQYRTRLCKDETG------CAR-RVCFFAHRRDELR 316
> At2g41900
Length=727
Score = 62.4 bits (150), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 46/129 (35%), Positives = 59/129 (45%), Gaps = 22/129 (17%)
Query 391 LSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDLDSSSLQ 445
+ SFKV PC +S HD CPF H RD R+F Y C D
Sbjct 275 MYSFKVRPCSRAYS--HDWTECPFVHPGENARRRDPRKF--HYSCVPCP-----DFRKGA 325
Query 446 CSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVRAPLF 505
C +GD C+ +H E HP+ ++ R C ++GT C R C FAHT EE+R PL+
Sbjct 326 CRRGDMCEYAHGVFECWLHPAQYRTRLC-----KDGTG-CAR-RVCFFAHTPEELR-PLY 377
Query 506 TVQEETCPS 514
PS
Sbjct 378 ASTGSAVPS 386
Score = 60.8 bits (146), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 42/142 (29%), Positives = 58/142 (40%), Gaps = 19/142 (13%)
Query 507 VQEETCPSSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYGSEPCPAWN 564
++ + +F M FK C HDW C + H ++ RR + Y PCP +
Sbjct 263 IKNSIYATDEFRMYSFKVRPCSRAYSHDWTECPFVHPGENARRRDPRKFHYSCVPCPDFR 322
Query 565 KDVHSADYSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAF 624
K C G +C Y+HG E HP+ Y+T C D G C R V C F
Sbjct 323 KGA--------CRRGDMCEYAHGVFECWLHPAQYRTRLCKD-------GTGCARRV-CFF 366
Query 625 YHEASERRWPVRMAIDYSSPLP 646
H E R P+ + + P P
Sbjct 367 AHTPEELR-PLYASTGSAVPSP 387
> At5g12850
Length=706
Score = 60.8 bits (146), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 45/149 (30%), Positives = 61/149 (40%), Gaps = 20/149 (13%)
Query 488 GHFCAFAHTREE--VRAPLFTVQEETCPSSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQ 545
G FA ++E + L ++ + +F M FK C HDW C +AH +
Sbjct 227 GTDVTFASEKKEYPIDPSLPDIKSGIYSTDEFRMFSFKIRPCSRAYSHDWTECPFAHPGE 286
Query 546 DCRRSP--QVGYGSEPCPAWNKDVHSADYSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPC 603
+ RR + Y PCP + K C G +C Y+HG E HP+ Y+T C
Sbjct 287 NARRRDPRKFHYTCVPCPDFKKG--------SCKQGDMCEYAHGVFECWLHPAQYRTRLC 338
Query 604 MDYRSKDRSGNSCPRGVLCAFYHEASERR 632
D G C R V C F H E R
Sbjct 339 KD-------GMGCNRRV-CFFAHANEELR 359
Score = 56.2 bits (134), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 58/129 (44%), Gaps = 22/129 (17%)
Query 391 LSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDLDSSSLQ 445
+ SFK+ PC +S HD CPF H RD R+F T C D S
Sbjct 260 MFSFKIRPCSRAYS--HDWTECPFAHPGENARRRDPRKFHYT-----CVPCPDFKKGS-- 310
Query 446 CSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVRAPLF 505
C +GD C+ +H E HP+ ++ R C ++G C R C FAH EE+R PL+
Sbjct 311 CKQGDMCEYAHGVFECWLHPAQYRTRLC-----KDGMG-CNR-RVCFFAHANEELR-PLY 362
Query 506 TVQEETCPS 514
PS
Sbjct 363 PSTGSGLPS 371
> At2g19810
Length=359
Score = 59.7 bits (143), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 44/136 (32%), Positives = 55/136 (40%), Gaps = 20/136 (14%)
Query 499 EVRAPLFTVQEETCPSSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYG 556
++ P + TC F M FK C G HDW C YAH + RR + Y
Sbjct 63 DLSGPDSPIDAYTC--DHFRMYEFKVRRCARGRSHDWTECPYAHPGEKARRRDPRKFHYS 120
Query 557 SEPCPAWNKDVHSADYSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSC 616
CP + K C G C +SHG E HP+ Y+T PC D G +C
Sbjct 121 GTACPEFRKGC--------CKRGDACEFSHGVFECWLHPARYRTQPCKD-------GGNC 165
Query 617 PRGVLCAFYHEASERR 632
R V C F H + R
Sbjct 166 RRRV-CFFAHSPDQIR 180
Score = 45.8 bits (107), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 53/202 (26%), Positives = 74/202 (36%), Gaps = 55/202 (27%)
Query 330 SHLDSSDTMHVAKISRDEPPSPAGSNSSGSGKGVEEIAVTPSGGKSTRLAF--------- 380
SH + T+H+ PP P + + V +I +P GG S A
Sbjct 6 SHRGFNPTVHI-------PPWPLSEDLT-----VSDIYGSPDGGSSMMEALAELQRYLPS 53
Query 381 NWPDSDGSLNLSS----FKVFPCRH-----------KHSVSHDKKYCPFYH-----NFRD 420
N PD D +LS + C H SHD CP+ H RD
Sbjct 54 NEPDPDSDPDLSGPDSPIDAYTCDHFRMYEFKVRRCARGRSHDWTECPYAHPGEKARRRD 113
Query 421 KRRFPVTYRAEQCEEHFDLDSSSLQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRN 480
R+F Y C E C +GDAC+ SH E HP+ ++ + C
Sbjct 114 PRKF--HYSGTACPEF-----RKGCCKRGDACEFSHGVFECWLHPARYRTQPCKDGGN-- 164
Query 481 GTERCGRGHFCAFAHTREEVRA 502
C R C FAH+ +++R
Sbjct 165 ----C-RRRVCFFAHSPDQIRV 181
> At2g25900
Length=315
Score = 58.9 bits (141), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 51/121 (42%), Gaps = 18/121 (14%)
Query 514 SSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYGSEPCPAWNKDVHSAD 571
S +F + FK C G HDW C +AH + RR + Y CP + K
Sbjct 88 SDEFRIYEFKIRRCARGRSHDWTECPFAHPGEKARRRDPRKFHYSGTACPEFRKG----- 142
Query 572 YSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASER 631
C G C +SHG E HPS Y+T PC D G SC R + C F H +
Sbjct 143 ---SCRRGDSCEFSHGVFECWLHPSRYRTQPCKD-------GTSCRRRI-CFFAHTTEQL 191
Query 632 R 632
R
Sbjct 192 R 192
Score = 46.6 bits (109), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 52/123 (42%), Gaps = 21/123 (17%)
Query 385 SDGSLNLSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDL 439
S + FK+ C SHD CPF H RD R+F Y C E
Sbjct 87 SSDEFRIYEFKIRRC--ARGRSHDWTECPFAHPGEKARRRDPRKF--HYSGTACPEF--- 139
Query 440 DSSSLQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREE 499
C +GD+C+ SH E HPS ++ + C ++GT R C FAHT E+
Sbjct 140 --RKGSCRRGDSCEFSHGVFECWLHPSRYRTQPC-----KDGTS--CRRRICFFAHTTEQ 190
Query 500 VRA 502
+R
Sbjct 191 LRV 193
> At4g29190
Length=356
Score = 58.5 bits (140), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 48/146 (32%), Positives = 63/146 (43%), Gaps = 27/146 (18%)
Query 517 FFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYGSEPCPAWNKDVHSADYSR 574
F M FK C G HDW C YAH + RR + Y CP + K
Sbjct 80 FRMYDFKVRRCARGRSHDWTECPYAHPGEKARRRDPRKYHYSGTACPDFRKG-------- 131
Query 575 RCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASERRW- 633
C G C ++HG E HP+ Y+T PC D G +C R + C F H + R+
Sbjct 132 GCKKGDSCEFAHGVFECWLHPARYRTQPCKD-------GGNCLRKI-CFFAHSPDQLRFL 183
Query 634 ----PVRM-AIDYSSPLPPQRSAALQ 654
P R+ + D SSP+ R+ A Q
Sbjct 184 HTRSPDRVDSFDVSSPI---RARAFQ 206
Score = 37.7 bits (86), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 51/118 (43%), Gaps = 21/118 (17%)
Query 389 LNLSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDLDSSS 443
+ FKV C S HD CP+ H RD R++ Y C D
Sbjct 80 FRMYDFKVRRCARGRS--HDWTECPYAHPGEKARRRDPRKY--HYSGTACP-----DFRK 130
Query 444 LQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVR 501
C KGD+C+ +H E HP+ ++ + C ++G C R C FAH+ +++R
Sbjct 131 GGCKKGDSCEFAHGVFECWLHPARYRTQPC-----KDGGN-CLR-KICFFAHSPDQLR 181
Score = 34.3 bits (77), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 20/36 (55%), Gaps = 2/36 (5%)
Query 112 CYSFDTGGANCASGLSCIFCHTAEELLLHPARFRSE 147
C F GG C G SC F H E LHPAR+R++
Sbjct 125 CPDFRKGG--CKKGDSCEFAHGVFECWLHPARYRTQ 158
> At5g58620
Length=607
Score = 56.2 bits (134), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 41/129 (31%), Positives = 55/129 (42%), Gaps = 22/129 (17%)
Query 391 LSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDLDSSSLQ 445
+ +FK+ PC +S HD CPF H RD R++ Y C E
Sbjct 212 MYAFKIKPCSRAYS--HDWTECPFVHPGENARRRDPRKY--HYSCVPCPEF-----RKGS 262
Query 446 CSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVRAPLF 505
CS+GD C+ +H E HP+ ++ R C C R C FAH EE+R PL+
Sbjct 263 CSRGDTCEYAHGIFECWLHPAQYRTRLCKDETN------CSR-RVCFFAHKPEELR-PLY 314
Query 506 TVQEETCPS 514
PS
Sbjct 315 PSTGSGVPS 323
Score = 55.5 bits (132), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 50/121 (41%), Gaps = 18/121 (14%)
Query 514 SSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYGSEPCPAWNKDVHSAD 571
+ +F M FK C HDW C + H ++ RR + Y PCP + K
Sbjct 207 TDEFRMYAFKIKPCSRAYSHDWTECPFVHPGENARRRDPRKYHYSCVPCPEFRKG----- 261
Query 572 YSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASER 631
C G C Y+HG E HP+ Y+T C D +C R V C F H+ E
Sbjct 262 ---SCSRGDTCEYAHGIFECWLHPAQYRTRLCKDE-------TNCSRRV-CFFAHKPEEL 310
Query 632 R 632
R
Sbjct 311 R 311
> At5g07500
Length=245
Score = 54.3 bits (129), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/136 (30%), Positives = 57/136 (41%), Gaps = 18/136 (13%)
Query 499 EVRAPLFTVQEETCPSSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDC-RRSP-QVGYG 556
E+ + + + S +F M +K CP HDW C YAH + RR P + Y
Sbjct 35 EIDPSIPNIDDAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTYC 94
Query 557 SEPCPAWNKDVHSADYSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSC 616
+ CPA+ C G C ++HG E HP+ Y+T C +GN C
Sbjct 95 AVACPAFRNGA--------CHRGDSCEFAHGVFEYWLHPARYRTRAC-------NAGNLC 139
Query 617 PRGVLCAFYHEASERR 632
R V C F H + R
Sbjct 140 QRKV-CFFAHAPEQLR 154
Score = 42.4 bits (98), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 52/118 (44%), Gaps = 21/118 (17%)
Query 389 LNLSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHFDLDSSS 443
+ ++K+ C + SHD CP+ H RD RR+ TY A C +
Sbjct 53 FRMYAYKIKRC--PRTRSHDWTECPYAHRGEKATRRDPRRY--TYCAVACPAF-----RN 103
Query 444 LQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTREEVR 501
C +GD+C+ +H E HP+ ++ R C N C R C FAH E++R
Sbjct 104 GACHRGDSCEFAHGVFEYWLHPARYRTRAC------NAGNLCQR-KVCFFAHAPEQLR 154
Score = 32.0 bits (71), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 2/43 (4%)
Query 104 RLLYWDRRCYSFDTGGANCASGLSCIFCHTAEELLLHPARFRS 146
R Y C +F G C G SC F H E LHPAR+R+
Sbjct 90 RYTYCAVACPAFRNGA--CHRGDSCEFAHGVFEYWLHPARYRT 130
> At1g03790
Length=393
Score = 50.4 bits (119), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 49/121 (40%), Gaps = 17/121 (14%)
Query 514 SSDFFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYGSEPCPAWNKDVHSAD 571
S F M FK C HDW C +AH + RR + Y E CP + + D
Sbjct 80 SDHFRMFEFKIRRCTRSRSHDWTDCPFAHPGEKARRRDPRRFQYSGEVCPEFRR---GGD 136
Query 572 YSRRCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASER 631
SR G C ++HG E HP Y+T C D G C R V C F H +
Sbjct 137 CSR----GDDCEFAHGVFECWLHPIRYRTEACKD-------GKHCKRKV-CFFAHSPRQL 184
Query 632 R 632
R
Sbjct 185 R 185
Score = 42.4 bits (98), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 40/127 (31%), Positives = 55/127 (43%), Gaps = 21/127 (16%)
Query 384 DSDGSLNLSSFKVFPCRHKH---SVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEE 435
DSD F++F + + S SHD CPF H RD RRF Y E C E
Sbjct 73 DSDDPYASDHFRMFEFKIRRCTRSRSHDWTDCPFAHPGEKARRRDPRRF--QYSGEVCPE 130
Query 436 HFDLDSSSLQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAH 495
CS+GD C+ +H E HP ++ C ++G + C R C FAH
Sbjct 131 F----RRGGDCSRGDDCEFAHGVFECWLHPIRYRTEAC-----KDG-KHCKRK-VCFFAH 179
Query 496 TREEVRA 502
+ ++R
Sbjct 180 SPRQLRV 186
Score = 36.2 bits (82), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 39/87 (44%), Gaps = 17/87 (19%)
Query 93 RRRPPVDEATGRLLYWDRRCYSFDTGGANCASGLSCIFCHTAEELLLHPARFRSELITSS 152
RRR P R Y C F GG +C+ G C F H E LHP R+R+E +
Sbjct 114 RRRDP-----RRFQYSGEVCPEFRRGG-DCSRGDDCEFAHGVFECWLHPIRYRTE----A 163
Query 153 TENAWHC-------ATSIAELRLRAPE 172
++ HC A S +LR+ PE
Sbjct 164 CKDGKHCKRKVCFFAHSPRQLRVLPPE 190
> At5g44260
Length=381
Score = 49.7 bits (117), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 39/118 (33%), Positives = 50/118 (42%), Gaps = 17/118 (14%)
Query 517 FFMQHFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP--QVGYGSEPCPAWNKDVHSADYSR 574
F M FK C HDW C ++H + RR + Y E CP +++ D SR
Sbjct 61 FRMYEFKIRRCTRSRSHDWTDCPFSHPGEKARRRDPRRFHYTGEVCPEFSR---HGDCSR 117
Query 575 RCPHGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASERR 632
G C ++HG E HPS Y+T C D G C R V C F H + R
Sbjct 118 ----GDECGFAHGVFECWLHPSRYRTEACKD-------GKHCKRKV-CFFAHSPRQLR 163
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 54/125 (43%), Gaps = 20/125 (16%)
Query 383 PDSDGSLNLSSFKVFPCRHKHSVSHDKKYCPFYH-----NFRDKRRFPVTYRAEQCEEHF 437
P + + FK+ C S SHD CPF H RD RRF Y E C E
Sbjct 55 PYAGDHFRMYEFKIRRC--TRSRSHDWTDCPFSHPGEKARRRDPRRF--HYTGEVCPEF- 109
Query 438 DLDSSSLQCSKGDACDKSHNRHELLYHPSIFKQRFCSSYATRNGTERCGRGHFCAFAHTR 497
S CS+GD C +H E HPS ++ C ++G + C R C FAH+
Sbjct 110 ---SRHGDCSRGDECGFAHGVFECWLHPSRYRTEAC-----KDG-KHCKRK-VCFFAHSP 159
Query 498 EEVRA 502
++R
Sbjct 160 RQLRV 164
Score = 32.3 bits (72), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 32/71 (45%), Gaps = 10/71 (14%)
Query 93 RRRPPVDEATGRLLYWDRRCYSFDTGGANCASGLSCIFCHTAEELLLHPARFRSELITSS 152
RRR P R Y C F G +C+ G C F H E LHP+R+R+E +
Sbjct 92 RRRDP-----RRFHYTGEVCPEFSRHG-DCSRGDECGFAHGVFECWLHPSRYRTE----A 141
Query 153 TENAWHCATSI 163
++ HC +
Sbjct 142 CKDGKHCKRKV 152
> Hs15281150
Length=756
Score = 38.1 bits (87), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 29/60 (48%), Gaps = 5/60 (8%)
Query 579 GTLCPYSH---GSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYHEASERRWPV 635
G CPY H G E+ YH YYKT C+ D G+ G+ CAF H + R PV
Sbjct 181 GFRCPYLHRTTGDTERKYHLRYYKTGTCI--HETDARGHCVKNGLHCAFAHGPLDLRPPV 238
Score = 35.8 bits (81), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 35/85 (41%), Gaps = 4/85 (4%)
Query 385 SDGSLNLSSFKVFPCRHKHSVSHDKKYCPFYHN--FRDKRRFPVTYRAEQCE--EHFDLD 440
SD + L S+K C + CP YHN R + YR+ C +H D
Sbjct 302 SDANFVLGSYKTEQCPKPPRLCRQGYACPHYHNSRDRRRNPRRFQYRSTPCPSVKHGDEW 361
Query 441 SSSLQCSKGDACDKSHNRHELLYHP 465
+C GD C H+R E +HP
Sbjct 362 GEPSRCDGGDGCQYCHSRTEQQFHP 386
Score = 34.7 bits (78), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 56/229 (24%), Positives = 84/229 (36%), Gaps = 50/229 (21%)
Query 417 NFRDKRRFPVTYRAEQCEEHFDLDSSSLQCSKGDACDKSHNRHELLYHPSIFKQRFCSSY 476
+F D +Y+ EQC + L C +G AC HN + +P F+ R
Sbjct 300 HFSDANFVLGSYKTEQCPKPPRL------CRQGYACPHYHNSRDRRRNPRRFQYRSTPCP 353
Query 477 ATRNGTE-----RCGRGHFCAFAHTREE--------VRAPL---FTVQEETCPSSDFFMQ 520
+ ++G E RC G C + H+R E V +P+ F+V +D
Sbjct 354 SVKHGDEWGEPSRCDGGDGCQYCHSRTEQQFHPEGPVGSPVSGAFSVFSHAGLVAD---- 409
Query 521 HFKTVWCPYGVQHDWYTCVYAHTYQDCRRSP-QVGYGSEPCPAWNKDV--HSADYSRRCP 577
++W Y + W+ ++ T P ++G E N AD + P
Sbjct 410 --PSLW--YPAEWSWHHVGHSRTINPEGDKPSRLGPAPENIKRGNDFACDGRADAAGMAP 465
Query 578 HGTLCPYSHGSKEQLYHPSYYKTMPCMDYRSKDRSGNSCPRGVLCAFYH 626
H + P YK+ C D R CPRG CAF H
Sbjct 466 HVCVFP-------------IYKSTKCNDMRQT----GYCPRGPFCAFAH 497
Lambda K H
0.315 0.128 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 26998981540
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40