bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1167_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
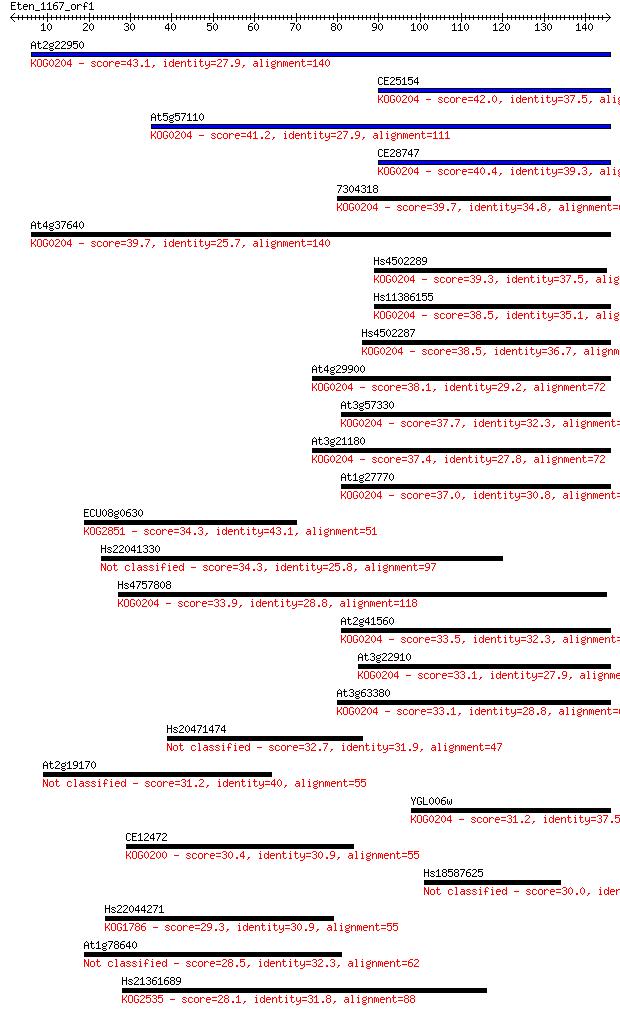
At2g22950 43.1 2e-04
CE25154 42.0 4e-04
At5g57110 41.2 7e-04
CE28747 40.4 0.001
7304318 39.7 0.002
At4g37640 39.7 0.002
Hs4502289 39.3 0.003
Hs11386155 38.5 0.004
Hs4502287 38.5 0.005
At4g29900 38.1 0.005
At3g57330 37.7 0.007
At3g21180 37.4 0.010
At1g27770 37.0 0.013
ECU08g0630 34.3 0.077
Hs22041330 34.3 0.091
Hs4757808 33.9 0.12
At2g41560 33.5 0.15
At3g22910 33.1 0.17
At3g63380 33.1 0.19
Hs20471474 32.7 0.25
At2g19170 31.2 0.62
YGL006w 31.2 0.78
CE12472 30.4 1.2
Hs18587625 30.0 1.6
Hs22044271 29.3 2.5
At1g78640 28.5 4.4
Hs21361689 28.1 6.6
> At2g22950
Length=1015
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 39/156 (25%), Positives = 69/156 (44%), Gaps = 23/156 (14%)
Query 6 LCCAVDSYEERRQLSPQLTRTYYSRRLSRRDSSVEGSFGTVEEGLLEHIS---------P 56
LC V + + R + + L++ Y + + R + V + + IS P
Sbjct 27 LCSVVKNPKRRFRFTANLSKRYEAAAMRRTNQEKLRIAVLVSKAAFQFISGVSPSDYKVP 86
Query 57 EQAE----DI-REKLTGLVKAYVTRTQCEDLTMLKNMGGADGIAHMLHVQRSVGLS--DP 109
E+ + DI ++L +V+ + D+ LK GG DG++ L + GLS +P
Sbjct 87 EEVKAAGFDICADELGSIVEGH-------DVKKLKFHGGVDGLSGKLKACPNAGLSTGEP 139
Query 110 QDIEHRRKLFGVNRLPSRASASFLSLCIDAASDNTL 145
+ + R++LFG+N+ SF +A D TL
Sbjct 140 EQLSKRQELFGINKFAESELRSFWVFVWEALQDMTL 175
> CE25154
Length=1228
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query 90 GADGIAHMLHVQRSVGLSDPQ-DIEHRRKLFGVNRLPSRASASFLSLCIDAASDNTL 145
G +G+ L VGL+ Q D++ RR ++G N +P S F+ L +DA D TL
Sbjct 44 GVEGLCKKLKTDSLVGLNGEQADLDRRRHVYGANTIPPAKSKGFVRLVLDACKDPTL 100
> At5g57110
Length=1099
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/112 (27%), Positives = 47/112 (41%), Gaps = 15/112 (13%)
Query 35 RDSSVEGSFGTVEEGLLEHISPEQAEDIREKLTGLVKAYVTRTQCEDLTMLKNMGGADGI 94
R+S VE + G I+PEQ V ++ + L+ GG G+
Sbjct 97 RESGVEKTTGPATPAGDFGITPEQ--------------LVIMSKDHNSGALEQYGGTQGL 142
Query 95 AHMLHVQRSVGLS-DPQDIEHRRKLFGVNRLPSRASASFLSLCIDAASDNTL 145
A++L G+S D D+ R+ ++G N P + FL DA D TL
Sbjct 143 ANLLKTNPEKGISGDDDDLLKRKTIYGSNTYPRKKGKGFLRFLWDACHDLTL 194
> CE28747
Length=1158
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query 90 GADGIAHMLHVQRSVGL-SDPQDIEHRRKLFGVNRLPSRASASFLSLCIDAASDNTL 145
G +G+ L GL +D +++EHRR FG N +P S SF L +A D TL
Sbjct 35 GVEGLCRKLKTDPINGLPNDTKELEHRRTAFGKNEIPPAPSKSFFRLAWEALQDITL 91
> 7304318
Length=999
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 35/67 (52%), Gaps = 1/67 (1%)
Query 80 EDLTMLKNMGGADGIAHMLHVQRSVGLSDPQ-DIEHRRKLFGVNRLPSRASASFLSLCID 138
E + + GG + L+ + GLS + D EHRR+ FG N +P + +FL+L +
Sbjct 28 EGVMKIAENGGIHELCKKLYTSPNEGLSGSKADEEHRRETFGSNVIPPKPPKTFLTLVWE 87
Query 139 AASDNTL 145
A D TL
Sbjct 88 ALQDVTL 94
> At4g37640
Length=1014
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 36/155 (23%), Positives = 66/155 (42%), Gaps = 22/155 (14%)
Query 6 LCCAVDSYEERRQLSPQLTRTYYS---RRLSRRDSSVEGSFGTVEEGLLEHISPEQ---A 59
LC V + + R + + L++ Y + RR ++ + + +SP
Sbjct 27 LCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQEKLRIAVLVSKAAFQFISGVSPSDYTVP 86
Query 60 EDIR--------EKLTGLVKAYVTRTQCEDLTMLKNMGGADGIAHMLHVQRSVGLS-DPQ 110
ED++ ++L +V+++ D+ LK GG DG+A L + GLS +
Sbjct 87 EDVKAAGFEICADELGSIVESH-------DVKKLKFHGGVDGLAGKLKASPTDGLSTEAA 139
Query 111 DIEHRRKLFGVNRLPSRASASFLSLCIDAASDNTL 145
+ R++LFG+N+ F +A D TL
Sbjct 140 QLSQRQELFGINKFAESEMRGFWVFVWEALQDMTL 174
> Hs4502289
Length=1205
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query 89 GGADGIAHMLHVQRSVGLS-DPQDIEHRRKLFGVNRLPSRASASFLSLCIDAASDNT 144
GG + L GLS +P D+E RR++FG N +P + +FL L +A D T
Sbjct 49 GGVQNLCSRLKTSPVEGLSGNPADLEKRRQVFGHNVIPPKKPKTFLELVWEALQDVT 105
> Hs11386155
Length=1173
Score = 38.5 bits (88), Expect = 0.004, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 1/58 (1%)
Query 89 GGADGIAHMLHVQRSVGLSD-PQDIEHRRKLFGVNRLPSRASASFLSLCIDAASDNTL 145
G G+ L + GL+D D+E RR+++G N +P + +FL L +A D TL
Sbjct 54 GDVSGLCRRLKTSPTEGLADNTNDLEKRRQIYGQNFIPPKQPKTFLQLVWEALQDVTL 111
> Hs4502287
Length=1220
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Query 86 KNMGGADGIAHMLHVQRSVGLS-DPQDIEHRRKLFGVNRLPSRASASFLSLCIDAASDNT 144
++ G GI L + GLS +P D+E R +FG N +P + +FL L +A D T
Sbjct 51 ESYGDVYGICTKLKTSPNEGLSGNPADLERREAVFGKNFIPPKKPKTFLQLVWEALQDVT 110
Query 145 L 145
L
Sbjct 111 L 111
> At4g29900
Length=1069
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query 74 VTRTQCEDLTMLKNMGGADGIAHMLHVQRSVGL-SDPQDIEHRRKLFGVNRLPSRASASF 132
V+ ++ +++ L+ +GG G++ +L G+ D DI R+ FG N P + SF
Sbjct 122 VSISRDQNIGALQELGGVRGLSDLLKTNLEKGIHGDDDDILKRKSAFGSNTYPQKKGRSF 181
Query 133 LSLCIDAASDNTL 145
+A+ D TL
Sbjct 182 WRFVWEASQDLTL 194
> At3g57330
Length=1025
Score = 37.7 bits (86), Expect = 0.007, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Query 81 DLTMLKNMGGADGIAHMLHVQRSVGLSDPQDIEHRRKLFGVNRLPSRASASFLSLCIDAA 140
D L +GG +GIA + V + G+ ++ R K++G NR + + SFL+ +A
Sbjct 108 DTKSLTKIGGPEGIAQKVSVSLAEGVRS-SELHIREKIYGENRYTEKPARSFLTFVWEAL 166
Query 141 SDNTL 145
D TL
Sbjct 167 QDITL 171
> At3g21180
Length=1090
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query 74 VTRTQCEDLTMLKNMGGADGIAHMLHVQRSVGLS-DPQDIEHRRKLFGVNRLPSRASASF 132
V+ T+ ++++ L+ GG G+A L G++ D +++ R+ FG N P + +F
Sbjct 136 VSMTRNQNMSNLQQYGGVKGVAEKLKSNMEQGINEDEKEVIDRKNAFGSNTYPKKKGKNF 195
Query 133 LSLCIDAASDNTL 145
+A D TL
Sbjct 196 FMFLWEAWQDLTL 208
> At1g27770
Length=1020
Score = 37.0 bits (84), Expect = 0.013, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 33/66 (50%), Gaps = 1/66 (1%)
Query 81 DLTMLKNMGGADGIAHMLHVQRSVGLSDPQD-IEHRRKLFGVNRLPSRASASFLSLCIDA 139
DL LK GG +G+ L + G+S +D + R++++G+N+ S F +A
Sbjct 111 DLKKLKIHGGTEGLTEKLSTSIASGISTSEDLLSVRKEIYGINQFTESPSRGFWLFVWEA 170
Query 140 ASDNTL 145
D TL
Sbjct 171 LQDTTL 176
> ECU08g0630
Length=298
Score = 34.3 bits (77), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 29/59 (49%), Gaps = 8/59 (13%)
Query 19 LSPQLTRTYYSR--------RLSRRDSSVEGSFGTVEEGLLEHISPEQAEDIREKLTGL 69
L PQL RTYY R R D S E SFG G + ++ A++ R+KL G+
Sbjct 20 LDPQLMRTYYRTLFPLDLMFRCLRIDESREISFGLGSGGYIRFLTFRTADEFRKKLMGI 78
> Hs22041330
Length=245
Score = 34.3 bits (77), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query 23 LTRTYYSRRLSRRDSSVEGSFGTVEEGLLEHISPEQAEDIREKLTGLVKAYVTRTQCEDL 82
+T ++ RR +++ +E G + SP+Q D L + ++VT++QC +
Sbjct 151 VTECFHERRAQLWSAALGNWLYLMEAGRIRSASPDQQPDPASSLCQIPVSFVTKSQCSSV 210
Query 83 TMLKNMGGADGIAHMLHVQ-RSVGLSDPQDIEHRRKLF 119
+L+++ A G ++ VQ ++VG P RK F
Sbjct 211 -VLRDL--ASGYTDVIQVQTQTVGEKVPLLENASRKHF 245
> Hs4757808
Length=1198
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 34/122 (27%), Positives = 49/122 (40%), Gaps = 27/122 (22%)
Query 27 YYSRRLSRRDSSVEGSFG-TVEE--GLLEHISPEQAEDIREKLTGLVKAYVTRTQCEDLT 83
+YS+ R +SS G FG T+EE L+E E I+E
Sbjct 9 FYSKN-QRNESSHGGEFGCTMEELRSLMELRGTEAVVKIKE------------------- 48
Query 84 MLKNMGGADGIAHMLHVQRSVGL-SDPQDIEHRRKLFGVNRLPSRASASFLSLCIDAASD 142
G + I L GL D+E R+++FG N +P + +FL L +A D
Sbjct 49 ---TYGDTEAICRRLKTSPVEGLPGTAPDLEKRKQIFGQNFIPPKKPKTFLQLVWEALQD 105
Query 143 NT 144
T
Sbjct 106 VT 107
> At2g41560
Length=1030
Score = 33.5 bits (75), Expect = 0.15, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 81 DLTMLKNMGGADGIAHMLHVQRSVGLSDPQDIEHRRKLFGVNRLPSRASASFLSLCIDAA 140
D L GG + +A + V S G+ ++ R K+FG NR + + SFL +A
Sbjct 108 DTKSLAQKGGVEELAKKVSVSLSEGIRS-SEVPIREKIFGENRYTEKPARSFLMFVWEAL 166
Query 141 SDNTL 145
D TL
Sbjct 167 HDITL 171
> At3g22910
Length=1017
Score = 33.1 bits (74), Expect = 0.17, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 31/62 (50%), Gaps = 1/62 (1%)
Query 85 LKNMGGADGIAHMLHVQRSVGLSDPQD-IEHRRKLFGVNRLPSRASASFLSLCIDAASDN 143
L+++GG +G+ L +G+++ D I+ RR FG N + S ++A D
Sbjct 100 LESLGGPNGLVSALKSNTRLGINEEGDEIQRRRSTFGSNTYTRQPSKGLFHFVVEAFKDL 159
Query 144 TL 145
T+
Sbjct 160 TI 161
> At3g63380
Length=1033
Score = 33.1 bits (74), Expect = 0.19, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query 80 EDLTMLKNMGGADGIAHMLHVQRSVGL-SDPQDIEHRRKLFGVNRLPSRASASFLSLCID 138
+DL ++ +GG +G+A L + G+ + Q++ RR LFG N L +
Sbjct 100 KDLPGIQALGGVEGVAASLRTNPTKGIHGNEQEVSRRRDLFGSNTYHKPPPKGLLFFVYE 159
Query 139 AASDNTL 145
A D T+
Sbjct 160 AFKDLTI 166
> Hs20471474
Length=370
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 39 VEGSFGTVEEGLLEHISPEQAEDIREKLTGLVKAYVTRTQCEDLTML 85
+E +FG + L ++P++ R TG+V YV+R + L ML
Sbjct 153 LEAAFGALHYFLFGFLAPKKFPCTRPPCTGVVDCYVSRPTEKSLLML 199
> At2g19170
Length=815
Score = 31.2 bits (69), Expect = 0.62, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 33/67 (49%), Gaps = 14/67 (20%)
Query 9 AVDSYEERRQLSPQLTRTYYSRRLSRRDSSVEG------------SFGTVEEGLLEHISP 56
AV+S +E+ S +L T Y+R L R+ + G S+ + G H+SP
Sbjct 45 AVES-DEKIDTSSELV-TVYARHLERKHDMILGMLFEEGSYKKLYSYKHLINGFAAHVSP 102
Query 57 EQAEDIR 63
EQAE +R
Sbjct 103 EQAETLR 109
> YGL006w
Length=1173
Score = 31.2 bits (69), Expect = 0.78, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 98 LHVQRSVGLSDPQDIEHRR----KLFGVNRLPSRASASFLSLCIDAASDNTL 145
L ++ G+S P+ +R+ K +G N LP R SFL L A +D T+
Sbjct 66 LKTDKNAGISLPEISNYRKTNRYKNYGDNSLPERIPKSFLQLVWAAFNDKTM 117
> CE12472
Length=495
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 29 SRRLSRRDSSVEGSFGTVEEGLLEHISPEQAEDIREKLTGLVKAYVTRTQCEDLT 83
S L + G+FG V +G+L+ SP+ E+ + +LT VK + C D++
Sbjct 137 SENLENKSILGSGNFGVVRKGILKMASPKNEEEKKMRLTVAVK---SAANCYDIS 188
> Hs18587625
Length=475
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 101 QRSVGLSDPQDIEHRRKLFGVNRLPSRASASFL 133
QR DP D+EH R+L + P A+ ++
Sbjct 247 QRVTSCHDPGDVEHSRRLISSPKFPVGATVQYI 279
> Hs22044271
Length=1513
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 24 TRTYYSRRLSRRDSSVEGSFGTVEEGLLEHISPEQAEDIREKLTGLVKAYVTRTQ 78
++ YY R S + V+ + EG PE ED K+ GLVK + R++
Sbjct 424 SKEYYMRLASGNPAIVQDAIVESSEGEAAQQEPEHGEDTIAKVKGLVKPPLKRSR 478
> At1g78640
Length=487
Score = 28.5 bits (62), Expect = 4.4, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 33/63 (52%), Gaps = 8/63 (12%)
Query 19 LSPQLTRTYYSRRLSRRDSSVEGSFGTVEEGLLEHISPEQAEDIREKLTGL-VKAYVTRT 77
+ +LT++ +RL+ SSVE E +L+H+ PE ++ I + G+ VK Y T
Sbjct 367 IKKELTKSDACQRLTLSKSSVE-------EHILKHLLPEDSQKIDKGKPGITVKVYDNDT 419
Query 78 QCE 80
E
Sbjct 420 DTE 422
> Hs21361689
Length=547
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 38/91 (41%), Gaps = 16/91 (17%)
Query 28 YSRRLSRRDSSVEGSFGTVEEGLLEHISPEQAEDIREKLTGLVKAYVTRTQCEDLTMLKN 87
Y L RRD G + E L + P+Q DI L GL++ +C + T
Sbjct 418 YQVELVRRDYVANGGW----ETFLSYEDPDQ--DI---LIGLLRL----RKCSEETFRFE 464
Query 88 MGGADGIAHMLHVQRS---VGLSDPQDIEHR 115
+GG I LHV S V DP +H+
Sbjct 465 LGGGVSIVRELHVYGSVVPVSSRDPTKFQHQ 495
Lambda K H
0.318 0.133 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40