bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1210_orf2
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
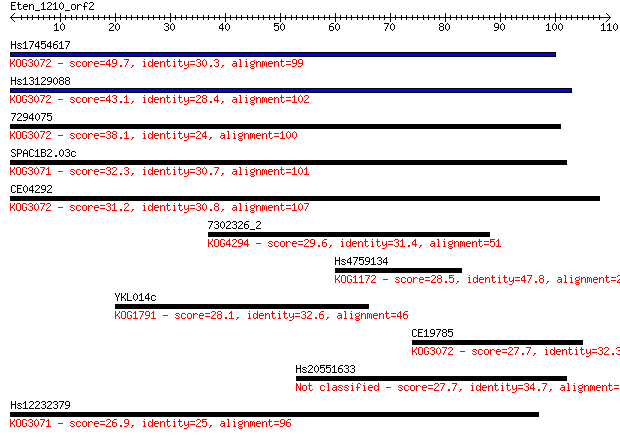
Hs17454617 49.7 1e-06
Hs13129088 43.1 1e-04
7294075 38.1 0.004
SPAC1B2.03c 32.3 0.23
CE04292 31.2 0.47
7302326_2 29.6 1.6
Hs4759134 28.5 3.4
YKL014c 28.1 3.7
CE19785 27.7 4.9
Hs20551633 27.7 5.7
Hs12232379 26.9 9.6
> Hs17454617
Length=270
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 52/99 (52%), Gaps = 17/99 (17%)
Query 1 YYFLAAQLRRPLSWGIFVTVAQISQMFVGVGVTFVSMYFAFSFPYRAFWRKEQIQDALSH 60
Y AA ++ P + +T QI QMFVG V+ ++ WR++Q
Sbjct 185 YTLKAANVKPPKMLPMLITSLQILQMFVGAIVSILT----------YIWRQDQ------- 227
Query 61 GHYITPKNLYWACLMYSTYFYLFAEYFVKRYVKGKEHSR 99
G + T ++L+W+ ++Y TYF LFA +F + Y++ K ++
Sbjct 228 GCHTTMEHLFWSFILYMTYFILFAHFFCQTYIRPKVKAK 266
> Hs13129088
Length=265
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 48/102 (47%), Gaps = 17/102 (16%)
Query 1 YYFLAAQLRRPLSWGIFVTVAQISQMFVGVGVTFVSMYFAFSFPYRAFWRKEQIQDALSH 60
Y AA R + +F+T++QI+QM +G V Y F + +Q H
Sbjct 181 YALRAAGFRVSRKFAMFITLSQITQMLMGCVVN----YLVFCW----------MQHDQCH 226
Query 61 GHYITPKNLYWACLMYSTYFYLFAEYFVKRYVKGKEHSRKAH 102
H+ +N++W+ LMY +Y LF +F + Y+ + KA
Sbjct 227 SHF---QNIFWSSLMYLSYLVLFCHFFFEAYIGKMRKTTKAE 265
> 7294075
Length=313
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/100 (24%), Positives = 43/100 (43%), Gaps = 14/100 (14%)
Query 1 YYFLAAQLRRPLSWGIFVTVAQISQMFVGVGVTFVSMYFAFSFPYRAFWRKEQIQDALSH 60
Y AA+ P + +T Q++QM +G + W ++ +
Sbjct 184 YALKAARFNPPRFISMIITSLQLAQMIIGCAINV--------------WANGFLKTHGTS 229
Query 61 GHYITPKNLYWACLMYSTYFYLFAEYFVKRYVKGKEHSRK 100
+I+ +N+ + MYS+YF LFA +F K Y+ H +
Sbjct 230 SCHISQRNINLSIAMYSSYFVLFARFFYKAYLAPGGHKSR 269
> SPAC1B2.03c
Length=334
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 50/103 (48%), Gaps = 10/103 (9%)
Query 1 YYFLAAQLRRPLSWGIFVTVAQISQMFVGVGVTFVSMYFAFSFPYRAFWRKEQIQDALSH 60
YYFLAA RR + W +VT QI Q + + + + Y +F Y F + D
Sbjct 204 YYFLAACGRR-VWWKQWVTRVQIIQFVLDLILCYFGTYSHIAFRY--FPWLPHVGDC--S 258
Query 61 GHYITPKNLYWACLMYSTYFYLFAEYFVKRYVK--GKEHSRKA 101
G ++ C + S+Y +LF +++ Y+K K++ RKA
Sbjct 259 GSLFAA---FFGCGVLSSYLFLFIGFYINTYIKRGAKKNQRKA 298
> CE04292
Length=435
Score = 31.2 bits (69), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 44/111 (39%), Gaps = 24/111 (21%)
Query 1 YYFLAAQLRRPLSWGIFVTVAQISQMFVGV--GVTFVSMYFAFSFPYRAFWRKEQIQDAL 58
Y + + R P + VT Q++QM +GV GVT YR
Sbjct 226 YALRSLKFRLPKQMAMVVTTLQLAQMVMGVIIGVTV----------YRI----------K 265
Query 59 SHGHYI--TPKNLYWACLMYSTYFYLFAEYFVKRYVKGKEHSRKAHKHCKH 107
S G Y T NL +Y TYF LFA +F YVK + + K+
Sbjct 266 SSGEYCQQTWDNLGLCFGVYFTYFLLFANFFYHAYVKKNNRTVNYENNSKN 316
> 7302326_2
Length=457
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 37 MYFAFSFPYRAFWRKEQIQDALSHGH-YITPKNLYWACLMYSTYFYLFAEYF 87
M S P + W E Q++ HG YI ++L+WA ++ T+ L + Y+
Sbjct 3 MESKMSRPKQPDWLAELCQESSIHGMPYIARRDLHWAERLFWTFIILGSAYY 54
> Hs4759134
Length=1044
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 1/23 (4%)
Query 60 HGHYITPKNLYWACLMYSTYFYL 82
HG Y TP L+W+C+++ T F L
Sbjct 687 HGPY-TPDVLFWSCILFFTTFIL 708
> YKL014c
Length=1764
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 22/46 (47%), Gaps = 3/46 (6%)
Query 20 VAQISQMFVGVGVTFVSMYFAFSFPYRAFWRKEQIQDALSHGHYIT 65
V + ++ F+ G S F+ +FP W K + D SHG IT
Sbjct 282 VKKANEFFLTFGA---SRDFSVAFPDNCVWFKNSVADGASHGAPIT 324
> CE19785
Length=274
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 10/31 (32%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 74 LMYSTYFYLFAEYFVKRYVKGKEHSRKAHKH 104
LMY +Y +LFA++F K Y++ + ++ + +
Sbjct 244 LMYISYLFLFAKFFYKAYIQKRSPTKTSKQE 274
> Hs20551633
Length=518
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 23/51 (45%), Gaps = 2/51 (3%)
Query 53 QIQDALSHGHYITP--KNLYWACLMYSTYFYLFAEYFVKRYVKGKEHSRKA 101
Q DA S + P KNL WA + TY +L + F ++K KA
Sbjct 164 QSSDADSEALFGVPNDKNLLWAYAVIGTYMFLVSVIFFCLFLKNSSKQEKA 214
> Hs12232379
Length=314
Score = 26.9 bits (58), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 24/100 (24%), Positives = 47/100 (47%), Gaps = 26/100 (26%)
Query 1 YYFLAA---QLRRPLSWGIFVTVAQISQMFVGVGVTFVSMYFAFSFPYRAFWRKEQIQDA 57
YY L A +++ L W ++T+ Q+ Q V +G T +S+Y F
Sbjct 198 YYGLTAFGPWIQKYLWWKRYLTMLQLIQFHVTIGHTALSLYTDCPF-------------- 243
Query 58 LSHGHYITPKNLYWACLMYS-TYFYLFAEYFVKRYVKGKE 96
PK ++WA + Y+ ++ +LF ++++ Y + K+
Sbjct 244 --------PKWMHWALIAYAISFIFLFLNFYIRTYKEPKK 275
Lambda K H
0.332 0.140 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1199474506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40