bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1233_orf3
Length=363
Score E
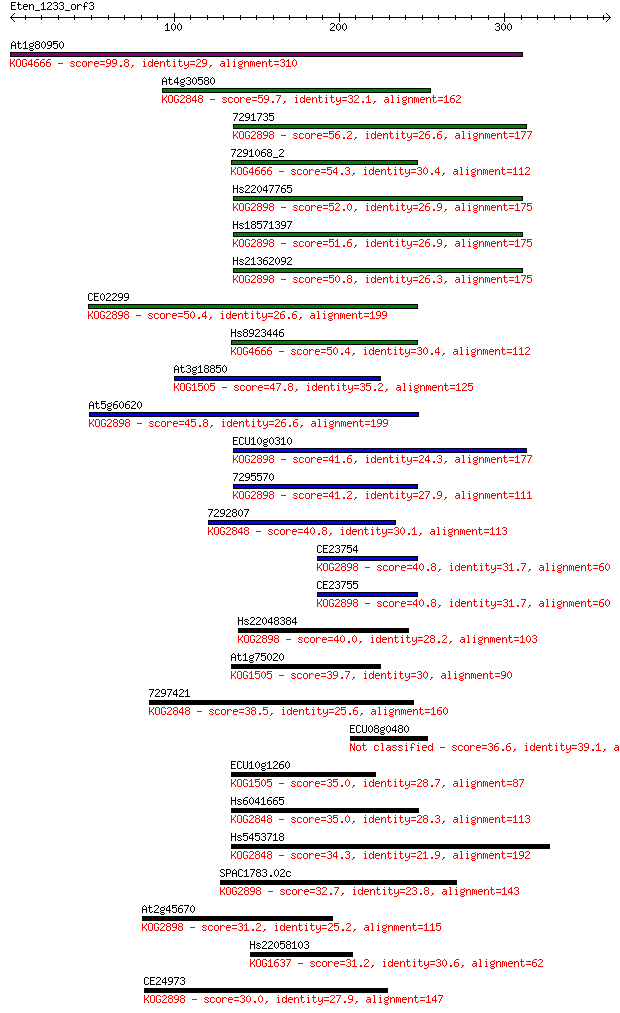
Sequences producing significant alignments: (Bits) Value
At1g80950 99.8 9e-21
At4g30580 59.7 1e-08
7291735 56.2 1e-07
7291068_2 54.3 4e-07
Hs22047765 52.0 2e-06
Hs18571397 51.6 2e-06
Hs21362092 50.8 5e-06
CE02299 50.4 5e-06
Hs8923446 50.4 6e-06
At3g18850 47.8 4e-05
At5g60620 45.8 1e-04
ECU10g0310 41.6 0.003
7295570 41.2 0.004
7292807 40.8 0.004
CE23754 40.8 0.004
CE23755 40.8 0.005
Hs22048384 40.0 0.007
At1g75020 39.7 0.011
7297421 38.5 0.022
ECU08g0480 36.6 0.084
ECU10g1260 35.0 0.27
Hs6041665 35.0 0.28
Hs5453718 34.3 0.39
SPAC1783.02c 32.7 1.2
At2g45670 31.2 3.4
Hs22058103 31.2 3.9
CE24973 30.0 7.6
> At1g80950
Length=398
Score = 99.8 bits (247), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 90/349 (25%), Positives = 146/349 (41%), Gaps = 56/349 (16%)
Query 1 DAAAALRWVRAAAAGWDRLTLQQQQQFAPFKRSDWHRLSTPKILLGAFFLAPLRFFLCVG 60
D AAA+ + A + R L PF ++ +L+ + L PLRF L +
Sbjct 29 DLAAAIEELDKKFAPYARTDLYGTMGLGPFPMTENIKLAVALVTL-----VPLRFLLSMS 83
Query 61 AMALHNMCIRAVALCFGTEEKLKKDQLLR--------------RRCLFAAISRFYCTIIL 106
+ L+ + R L ++++ +R + RF ++L
Sbjct 84 ILLLYYLICRVFTLFSAPYRGPEEEEDEGGVVFQEDYAHMEGWKRTVIVRSGRFLSRVLL 143
Query 107 RLFGVVSTRSFFW------------DGKERIGPDAFECKN------------IVSNHVSC 142
+FG F+W D + K IVSNHVS
Sbjct 144 FVFG------FYWIHESCPDRDSDMDSNPKTTSTEINQKGEAATEEPERPGAIVSNHVSY 197
Query 143 MDIMYFLSQAFTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRALEDLV-ERMKM 201
+DI+Y +S +F +FV K S+ + PL+ ++ GC+ V+R++ + + + V ER++
Sbjct 198 LDILYHMSASFPSFVAKRSVGKLPLVGLISKCLGCVYVQREAKSPDFKGVSGTVNERVRE 257
Query 202 NPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIYTWGCIHPAYELIPTP 261
S+ + I++FPEGTTTNG L+ F+ GAF A V P +L Y + A++ I
Sbjct 258 AHSNKSAPTIMLFPEGTTTNGDYLLTFKTGAFLAGTPVLPVILKYPYERFSVAWDTISGA 317
Query 262 YFISLSLSAWPASELRAYWLPPIDPPAPEGFDSEEKRIAAFATQVRNAM 310
I L L + L LP P E D + +A+ VR M
Sbjct 318 RHI-LFLLCQVVNHLEVIRLPVYYPSQEEKDDPK-----LYASNVRKLM 360
> At4g30580
Length=212
Score = 59.7 bits (143), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 52/170 (30%), Positives = 76/170 (44%), Gaps = 16/170 (9%)
Query 93 LFAAISRFYCTIILRLFGVVSTRSFF---WDGKERIGPDAFECKNIVSNHVSCMDIMYFL 149
LF R + I +L+ +S F+ +G E + P + VSNH S +DI L
Sbjct 10 LFDPYRRKFHHFIAKLWASISIYPFYKINIEGLENL-PSSDTPAVYVSNHQSFLDIYTLL 68
Query 150 SQAFT-AFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRALEDLVERMKMNPSDPAC 208
S + F+ K IF P+I W G +P+KR + L+ +E +K S
Sbjct 69 SLGKSFKFISKTGIFVIPIIGWAMSMMGVVPLKRMDPRSQVDCLKRCMELLKKGAS---- 124
Query 209 NPIVVFPEGTTTNGRCLIQFRRGAFTALQR----VQPCVLIYTWGCIHPA 254
+ FPEGT + L F++GAFT + V P L+ T G I P
Sbjct 125 --VFFFPEGTRSKDGRLGSFKKGAFTVAAKTGVAVVPITLMGT-GKIMPT 171
> 7291735
Length=537
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 47/185 (25%), Positives = 84/185 (45%), Gaps = 25/185 (13%)
Query 136 VSNHVSCMDIMYFLSQAFTAFVC-KASIFQTPLIAWPARFFGCIPVKRDSDADRHRALED 194
V+NH S +D++ + + + + + F L AR I +R DRH
Sbjct 331 VANHTSPIDVLVLMCDSTYSLIGQRHGGFLGVLQRALARASPHIWFERGEAKDRHL---- 386
Query 195 LVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIYT------- 247
+ ER+K + SDP PI++FPEGT N ++QF++G+F + P + Y
Sbjct 387 VAERLKQHVSDPNNPPILIFPEGTCINNTSVMQFKKGSFEVGGVIYPVAIKYDPRFGDAF 446
Query 248 WGCIHPAYELIPTPYFISLSLSAWPASELRAYWLPPIDPPAPEGFDSEEKRIAAFATQVR 307
W Y ++ Y + +++W A ++LPP+ + E + FA +V+
Sbjct 447 WNS--AKYSMMQYLYMM---MTSW-AIVCDVWYLPPM-------YRQEGESAIDFANRVK 493
Query 308 NAMWK 312
+ + K
Sbjct 494 SVIAK 498
> 7291068_2
Length=402
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 54/112 (48%), Gaps = 3/112 (2%)
Query 135 IVSNHVSCMDIMYFLSQAFTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRALED 194
+V+ H S +D + ++ + V K PL+ + I V+R+ R + D
Sbjct 84 VVAPHSSYVDSILVVASGPPSIVAKRETADIPLLGKIINYAQPIYVQREDPNSRQNTIRD 143
Query 195 LVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIY 246
+V+R + P +V+F EGT TN LI+F+ GAF VQP +L Y
Sbjct 144 IVDRARSTDDWPQ---VVIFAEGTCTNRTALIKFKPGAFYPGVPVQPVLLKY 192
> Hs22047765
Length=248
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 47/188 (25%), Positives = 84/188 (44%), Gaps = 35/188 (18%)
Query 136 VSNHVSCMDIMYFLSQAFTAFVCKA-----SIFQTPLI-AWPARFFGCIPVKRDSDADRH 189
V+NH S +D++ S + A V + + Q ++ A P +F +R DRH
Sbjct 37 VANHTSPIDVIILASDGYYAMVGQVHGGLMGVIQRAMVKACPHVWF-----ERSEVKDRH 91
Query 190 RALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIYT-- 247
+ L E ++ D + PI++FPEGT N ++ F++G+F V P + Y
Sbjct 92 LVAKRLTEHVQ----DKSKLPILIFPEGTCINNTSVMMFKKGSFEIGATVYPVAIKYDPQ 147
Query 248 -----WGCIHPAYELIPTPYFISLSLSAWPASELRAYWLPPIDPPAPEGFDSEEKRIAAF 302
W Y ++ Y + + +++W A ++LPP+ A E F
Sbjct 148 FGDAFWNS--SKYGMV--TYLLRM-MTSW-AIVCSVWYLPPMTREADED-------AVQF 194
Query 303 ATQVRNAM 310
A +V++A+
Sbjct 195 ANRVKSAI 202
> Hs18571397
Length=456
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 47/188 (25%), Positives = 84/188 (44%), Gaps = 35/188 (18%)
Query 136 VSNHVSCMDIMYFLSQAFTAFVCKA-----SIFQTPLI-AWPARFFGCIPVKRDSDADRH 189
V+NH S +D++ S + A V + + Q ++ A P +F +R DRH
Sbjct 245 VANHTSPIDVIILASDGYYAMVGQVHGGLMGVIQRAMVKACPHVWF-----ERSEVKDRH 299
Query 190 RALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIYT-- 247
+ L E ++ D + PI++FPEGT N ++ F++G+F V P + Y
Sbjct 300 LVAKRLTEHVQ----DKSKLPILIFPEGTCINNTSVMMFKKGSFEIGATVYPVAIKYDPQ 355
Query 248 -----WGCIHPAYELIPTPYFISLSLSAWPASELRAYWLPPIDPPAPEGFDSEEKRIAAF 302
W Y ++ Y + + +++W A ++LPP+ A E F
Sbjct 356 FGDAFWNS--SKYGMV--TYLLRM-MTSW-AIVCSVWYLPPMTREADED-------AVQF 402
Query 303 ATQVRNAM 310
A +V++A+
Sbjct 403 ANRVKSAI 410
> Hs21362092
Length=434
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 46/188 (24%), Positives = 87/188 (46%), Gaps = 35/188 (18%)
Query 136 VSNHVSCMDIMYFLSQAFTAFVCKA-----SIFQTPLI-AWPARFFGCIPVKRDSDADRH 189
V+NH S +D++ + A V + I Q ++ A P +F +R DRH
Sbjct 226 VANHTSPIDVLILTTDGCYAMVGQVHGGLMGIIQRAMVKACPHVWF-----ERSEMKDRH 280
Query 190 RALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIYT-- 247
+ +R+K + +D PI++FPEGT N ++ F++G+F + P + Y
Sbjct 281 L----VTKRLKEHIADKKKLPILIFPEGTCINNTSVMMFKKGSFEIGGTIHPVAIKYNPQ 336
Query 248 -----WGCIHPAYELIPTPYFISLSLSAWPASELRAYWLPPIDPPAPEGFDSEEKRIAAF 302
W Y ++ Y + + +++W A +++PP+ EG D+ + F
Sbjct 337 FGDAFWNS--SKYNMV--SYLLRM-MTSW-AIVCDVWYMPPM--TREEGEDAVQ-----F 383
Query 303 ATQVRNAM 310
A +V++A+
Sbjct 384 ANRVKSAI 391
> CE02299
Length=512
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 53/200 (26%), Positives = 84/200 (42%), Gaps = 21/200 (10%)
Query 48 FFLAPLRFFLCVGAMALHNMCIRAVALCFGTEEKLKKDQLLRRRCLFAAISRFYCTIILR 107
+ L P R L A+ L M + + K++K L RRC+ + R Y R
Sbjct 169 YVLVPCRIALFGIAIVL--MIVSTSIIGLVPNAKVRK--FLNRRCMLMCM-RIYS----R 219
Query 108 LFGVVSTRSFFWDGKERIGPDAFECKNIVSNHVSCMDIMYFLSQAFTAFVC-KASIFQTP 166
F V F D + R C V+NH S +D+M A + K + F
Sbjct 220 AFSSVIR---FHDKENRANKGGI-C---VANHTSPIDVMVLSCDNCYAMIGQKQAGFLGF 272
Query 167 LIAWPARFFGCIPVKRDSDADRHRALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLI 226
L +R I +R DR + ++ RM+ + +D PI++FPEGT N ++
Sbjct 273 LQTTLSRSEHHIWFERGEAGDRAKVMD----RMREHVNDENKLPIIIFPEGTCINNTSVM 328
Query 227 QFRRGAFTALQRVQPCVLIY 246
F++G+F + P + Y
Sbjct 329 MFKKGSFEIGSTIYPIAVKY 348
> Hs8923446
Length=427
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 50/112 (44%), Gaps = 3/112 (2%)
Query 135 IVSNHVSCMDIMYFLSQAFTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRALED 194
+ + H + D + + + V + Q PLI R + V R R + +
Sbjct 25 VAAPHSTFFDGIACVVAGLPSMVSRNENAQVPLIGRLLRAVQPVLVSRVDPDSRKNTINE 84
Query 195 LVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIY 246
+++R P I+VFPEGT TN CLI F+ GAF VQP +L Y
Sbjct 85 IIKRTTSGGEWPQ---ILVFPEGTCTNRSCLITFKPGAFIPGVPVQPVLLRY 133
> At3g18850
Length=375
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 44/154 (28%), Positives = 66/154 (42%), Gaps = 33/154 (21%)
Query 100 FYCTIILRLFGVVSTR---SFFWDGKERIGPDAFECKN------------------IVSN 138
F ++LRLF + +R SFF+ + P FE N +++N
Sbjct 40 FLSAVVLRLFSIRYSRKCVSFFFGSWLALWPFLFEKINKTKVIFSGDKVPCEDRVLLIAN 99
Query 139 HVSCMDIMYFLSQAF-------TAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRA 191
H + +D MYF A +V K+S+ + PL W F IPV+R + D
Sbjct 100 HRTEVDWMYFWDLALRKGQIGNIKYVLKSSLMKLPLFGWAFHLFEFIPVERRWEVDEAN- 158
Query 192 LEDLVERMKMNPSDPACNPIVVFPEGTT-TNGRC 224
L +V K +P D + +FPEGT T +C
Sbjct 159 LRQIVSSFK-DPRDALW--LALFPEGTDYTEAKC 189
> At5g60620
Length=376
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 53/201 (26%), Positives = 82/201 (40%), Gaps = 21/201 (10%)
Query 49 FLAPLR-FFLCVGAMALHNMCIRAVALCFGTEEKLKKDQLLRRRCLFAAISRFYCTIILR 107
L PLR F L G + ++ I AL G + KK + R L I F+ +
Sbjct 95 ILFPLRCFTLAFGWIIFLSLFIPVNALLKGQDRLRKKIE----RVLVEMICSFF---VAS 147
Query 108 LFGVVSTRSFFWDGKERIGPDAFECKNIVSNHVSCMDIMYFLSQ-AFTAFVCKASIFQTP 166
GVV + + I P + V+NH S +D + AF + K +
Sbjct 148 WTGVVK----YHGPRPSIRPK----QVYVANHTSMIDFIVLEQMTAFAVIMQKHPGWVGL 199
Query 167 LIAWPARFFGCIPVKRDSDADRHRALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLI 226
L + GCI R DR E + ++++ + NP+++FPEGT N +
Sbjct 200 LQSTILESVGCIWFNRSEAKDR----EIVAKKLRDHVQGADSNPLLIFPEGTCVNNNYTV 255
Query 227 QFRRGAFTALQRVQPCVLIYT 247
F++GAF V P + Y
Sbjct 256 MFKKGAFELDCTVCPIAIKYN 276
> ECU10g0310
Length=451
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 43/190 (22%), Positives = 74/190 (38%), Gaps = 25/190 (13%)
Query 136 VSNHVSCMDIMYFLSQAFTAFVCKAS--------IFQTPLIAWPARFFGCIPVKRDSDAD 187
V+NH S +D+ S F C + +F++ LI G I KR D
Sbjct 126 VANHTSFVDLFLLSSHRF-PHACVSERHGGLFGFLFKSILIRN-----GSIAFKRSEKID 179
Query 188 RHRALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIYT 247
R +E + E + + P+++FPEGT N + + F++G F V P + +
Sbjct 180 RQLVVEKVKEHVWSGGA-----PMLIFPEGTCVNNKFSVLFQKGPFELGVAVCPVAIRFQ 234
Query 248 WGCIHPAYELIPTPYFISL--SLSAWPASELRAYWLPPIDPPAPEG---FDSEEKRIAAF 302
P + + + + ++ W E W+ P+ E F K I +
Sbjct 235 RRLFDPYWNRRSHGFTMHMFYLMTRWRL-EAEITWMEPVRIMKDETSTQFSHRVKTIISK 293
Query 303 ATQVRNAMWK 312
+RN +W
Sbjct 294 EAGLRNTLWN 303
> 7295570
Length=407
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 31/116 (26%), Positives = 51/116 (43%), Gaps = 13/116 (11%)
Query 136 VSNHVSCMDIMYFLSQAFTAFVCKA-----SIFQTPLIAWPARFFGCIPVKRDSDADRHR 190
V NH S +D++ + A + + + Q L +R + R ADR
Sbjct 217 VCNHTSPLDVLVLMCDANYSLTGQVHTGILGVLQRAL----SRVSHHMWFDRKELADREA 272
Query 191 ALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIY 246
LV R+ + D P+++FPEGT N ++QF++G+F V P + Y
Sbjct 273 L--GLVLRLHCSMKDRP--PVLLFPEGTCINNTAVMQFKKGSFAVSDVVHPVAIRY 324
> 7292807
Length=343
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 48/117 (41%), Gaps = 11/117 (9%)
Query 121 GKERIGPDAFECKNIVSNHVSCMDIMYFLSQAFTAFVCKASIFQTPLIAWP----ARFFG 176
GKE + D + IV+NH S +D++ + C + AWP A G
Sbjct 85 GKEHLAKD--QACIIVANHQSSLDVLGMFNIWHVMNKCTVVAKRELFYAWPFGLAAWLAG 142
Query 177 CIPVKRDSDADRHRALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAF 233
I + R L D+ R+K + VFPEGT N L F++GAF
Sbjct 143 LIFIDRVRGEKARETLNDVNRRIKKQRIK-----LWVFPEGTRRNTGALHPFKKGAF 194
> CE23754
Length=617
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 187 DRHRALEDLV--ERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVL 244
+R A + LV +++K + ++P PI++FPEGT N ++ F++G+F + P +
Sbjct 380 ERSEAKDRLVVAQKLKEHCTNPDKLPILIFPEGTCINNTSVMMFKKGSFEIGTTIYPIAM 439
Query 245 IY 246
Y
Sbjct 440 KY 441
> CE23755
Length=407
Score = 40.8 bits (94), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 187 DRHRALEDLV--ERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVL 244
+R A + LV +++K + ++P PI++FPEGT N ++ F++G+F + P +
Sbjct 170 ERSEAKDRLVVAQKLKEHCTNPDKLPILIFPEGTCINNTSVMMFKKGSFEIGTTIYPIAM 229
Query 245 IY 246
Y
Sbjct 230 KY 231
> Hs22048384
Length=507
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 51/109 (46%), Gaps = 15/109 (13%)
Query 139 HVSCMDIMYFLSQAFTAFVCKA-----SIFQTPLIAW-PARFFGCIPVKRDSDADRHRAL 192
H S +D++ S + A V + + Q ++ P +F + VK DRH
Sbjct 332 HTSPIDVIILASDGYYATVGQMHGGLIGLIQRAMVKTCPHIWFERLEVK-----DRHLLA 386
Query 193 EDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQP 241
+ L E+++ D + PI++FPEG N ++ F++G+F V P
Sbjct 387 KRLTEQVQ----DISKLPILIFPEGICINNTSVMMFKKGSFEIGATVYP 431
> At1g75020
Length=393
Score = 39.7 bits (91), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 49/98 (50%), Gaps = 12/98 (12%)
Query 135 IVSNHVSCMDIMYFLSQA-------FTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDAD 187
+++NH + +D MY + A + +V K+S+ + P+ W IPV+R + D
Sbjct 101 LIANHRTEVDWMYLWNIALRKGCLGYIKYVLKSSLMKLPIFGWGFHVLEFIPVERKREVD 160
Query 188 RHRALEDLVERMKMNPSDPACNPIVVFPEGTT-TNGRC 224
L+ ++ K +P +P + +FPEGT T +C
Sbjct 161 EPVLLQ-MLSSFK-DPQEPLW--LALFPEGTDFTEEKC 194
> 7297421
Length=259
Score = 38.5 bits (88), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 41/168 (24%), Positives = 72/168 (42%), Gaps = 19/168 (11%)
Query 85 DQLLRRRCLFAAISRFYCTIILRLFGVVSTRSFFWDGKERIGPDAFECKNIVSNHVSCMD 144
D L R +A +R + RL G+ + G E + D ++ NH S +D
Sbjct 36 DSLSVRALYDSATARLSKCALCRLVGI----TMEVRGLENVRKDHGAV--VIMNHQSAVD 89
Query 145 --IMYFLSQAF--TAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRALEDLVERMK 200
++ +L V K + P A +G + + R D +L+ + ++
Sbjct 90 LCVLAYLWPVIGRATVVSKKEVLYIPFFGIGAWLWGTLYIDRSRKTDSINSLQKEAKAIQ 149
Query 201 MNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAF-TALQR---VQPCVL 244
+ C +++FPEGT + L+ F++G+F ALQ VQP V+
Sbjct 150 ----ERNCK-LLLFPEGTRNSKDSLLPFKKGSFHIALQGKSPVQPVVI 192
> ECU08g0480
Length=223
Score = 36.6 bits (83), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 4/46 (8%)
Query 207 ACNPIVVFPEGTTTNGRCLIQFRRGAFTALQRVQPCVLIYTWGCIH 252
A P V+FPEG TN + ++QF R ++ C + YT GCI+
Sbjct 123 ATEPCVLFPEGCQTNNKAVLQFSRD----VEVDHVCGIRYTGGCIN 164
> ECU10g1260
Length=357
Score = 35.0 bits (79), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 42/94 (44%), Gaps = 12/94 (12%)
Query 135 IVSNHVSCMDIMYF--LSQAFTAF-----VCKASIFQTPLIAWPARFFGCIPVKRDSDAD 187
IVSNH++ D ++ + +F F + K S+ P++ + RFF I + R D
Sbjct 88 IVSNHLTEYDWLFMCCVLDSFGRFENLCIILKMSLRDIPILGYGMRFFQFIFLNRKISKD 147
Query 188 RHRALEDLVERMKMNPSDPACNPIVVFPEGTTTN 221
R + K D +++FPEGT +
Sbjct 148 RELIISGTSRLKKKEKYD-----LLIFPEGTYID 176
> Hs6041665
Length=278
Score = 35.0 bits (79), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 52/118 (44%), Gaps = 11/118 (9%)
Query 135 IVSNHVSCMDIMYF---LSQAFTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRA 191
IVSNH S +D+M L + + +F P + G + R +
Sbjct 94 IVSNHQSILDMMGLMEVLPERCVQIAKRELLFLGP-VGLIMYLGGVFFINRQRSSTAMTV 152
Query 192 LEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAF-TALQRVQPCV-LIYT 247
+ DL ERM + ++PEGT + L+ F++GAF A+Q P V ++Y+
Sbjct 153 MADLGERMVRENLK-----VWIYPEGTRNDNGDLLPFKKGAFYLAVQAQVPIVPVVYS 205
> Hs5453718
Length=283
Score = 34.3 bits (77), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 42/195 (21%), Positives = 77/195 (39%), Gaps = 23/195 (11%)
Query 135 IVSNHVSCMDIMYFLS--QAFTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRAL 192
+VSNH S +D++ + + K + G I + R D +
Sbjct 100 VVSNHQSSLDLLGMMEVLPGRCVPIAKRELLWAGSAGLACWLAGVIFIDRKRTGDAISVM 159
Query 193 EDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAF-TALQRVQPCVLIYTWGCI 251
++ + + + VFPEGT + ++ F+RGAF A+Q P V I +
Sbjct 160 SEVAQTLLTQDVR-----VWVFPEGTRNHNGSMLPFKRGAFHLAVQAQVPIVPI-----V 209
Query 252 HPAYELIPTPYFISLSLSAWPASELRAYWLPPIDPPAPEGFDSEEKRIAAFATQVRNAMW 311
+Y+ F + + + + LPP+ EG ++ + A A +VR++M
Sbjct 210 MSSYQ-----DFYCKKERRFTSGQCQVRVLPPV---PTEGLTPDD--VPALADRVRHSML 259
Query 312 KVLKKEHFDAAEFSD 326
V ++ D D
Sbjct 260 TVFREISTDGRGGGD 274
> SPAC1783.02c
Length=328
Score = 32.7 bits (73), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 34/147 (23%), Positives = 61/147 (41%), Gaps = 14/147 (9%)
Query 128 DAFECKNIVS-NHVSCMDIMYFLSQAFTAF-VCKASIFQTPLIAWPARFFGCIPVKRDSD 185
D+ + +I++ NH S +D++ F VC + +I+ A F+ C
Sbjct 131 DSLQPGDILAVNHSSPLDVLVLSCLYNCTFAVCDSKTSNVSIISAQAYFWSCFFSPSKLK 190
Query 186 ADRHRALEDLVERMKMNPSDPACNPIVVFPEGTTTNGRCLIQFRRGAFTALQ--RVQPCV 243
+ L + + + +++FPEG TNGR L QF +A + R+ P
Sbjct 191 ITDAKPLAKVAAK-----ASKIGTVVILFPEGVCTNGRALCQFTPCFDSAKETDRIFPLY 245
Query 244 LIYTWGCIHPAYELIPTPYFISLSLSA 270
+ Y C+ +P P +S + S
Sbjct 246 IKYLPPCVT-----LPVPSLLSFARSV 267
> At2g45670
Length=281
Score = 31.2 bits (69), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 45/115 (39%), Gaps = 6/115 (5%)
Query 81 KLKKDQLLRRRCLFAAISRFYCTIILRLFGVVSTRSFFWDGKERIGPDAFECKNIVSNHV 140
K K++ + RC I+R IL FG R + I P +VSNHV
Sbjct 126 KDKENPMPLWRCRIMWITRICTRCILFSFGYQWIRRKGKPARREIAPI------VVSNHV 179
Query 141 SCMDIMYFLSQAFTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRALEDL 195
S ++ +++ + V S P + R I V R S R A+ ++
Sbjct 180 SYIEPIFYFYELSPTIVASESHDSLPFVGTIIRAMQVIYVNRFSQTSRKNAVHEI 234
> Hs22058103
Length=707
Score = 31.2 bits (69), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 27/62 (43%), Gaps = 3/62 (4%)
Query 146 MYFLSQAFTAFVCKASIFQTPLIAWPARFFGCIPVKRDSDADRHRALEDLVERMKMNPSD 205
+ FL ++ F FQ L P F G I + +++ +L D E KMNP D
Sbjct 464 LQFLQSVYSTF---GFSFQLNLSTRPENFLGEIEMWNEAEKQLQNSLMDFGEPWKMNPGD 520
Query 206 PA 207
A
Sbjct 521 GA 522
> CE24973
Length=390
Score = 30.0 bits (66), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 41/149 (27%), Positives = 62/149 (41%), Gaps = 33/149 (22%)
Query 82 LKKDQLLRRRCLFAAISRFYCTIILRLFGVVSTRSFFWDGKERIGPDAFECKNIVSNHVS 141
L+K LR L R C+I+ G+V +S D R+ + +NHVS
Sbjct 63 LRKSAALRMHTL-----RIMCSIL----GIVVEKSGHRDESARV---------LCANHVS 104
Query 142 CMDIMYFLSQAFTAFVCKASIFQTP-LIAWPARFFGCIPVKRDSDADRHRALEDLVERMK 200
+D L+ S++ P +I W FG + D R + LV R K
Sbjct 105 ILD---HLAVDILTPCLLPSVWDIPSIIRW---CFGYV------DLGATRGRDQLVSRAK 152
Query 201 MNPSDPACNPIVVFPEGTTTNG-RCLIQF 228
+ P++ FPEG T+G + LI+F
Sbjct 153 QLLTREQM-PLLAFPEGIITSGEKALIKF 180
Lambda K H
0.327 0.138 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8875059358
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40