bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
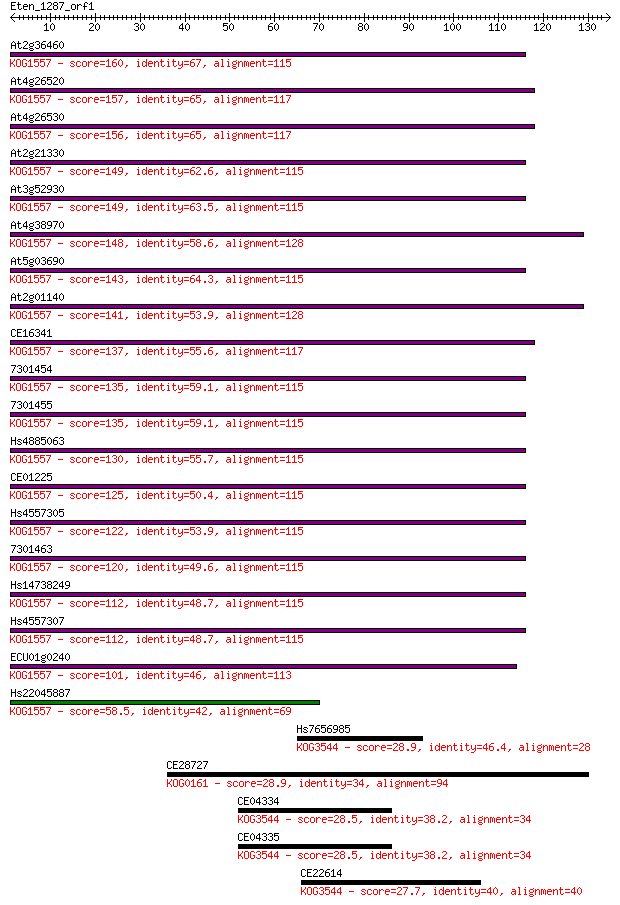
Query= Eten_1287_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
At2g36460 160 7e-40
At4g26520 157 6e-39
At4g26530 156 1e-38
At2g21330 149 1e-36
At3g52930 149 1e-36
At4g38970 148 3e-36
At5g03690 143 1e-34
At2g01140 141 3e-34
CE16341 137 7e-33
7301454 135 2e-32
7301455 135 3e-32
Hs4885063 130 7e-31
CE01225 125 3e-29
Hs4557305 122 1e-28
7301463 120 8e-28
Hs14738249 112 2e-25
Hs4557307 112 2e-25
ECU01g0240 101 3e-22
Hs22045887 58.5 3e-09
Hs7656985 28.9 2.9
CE28727 28.9 3.2
CE04334 28.5 4.0
CE04335 28.5 4.2
CE22614 27.7 6.2
> At2g36460
Length=358
Score = 160 bits (405), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 77/115 (66%), Positives = 89/115 (77%), Gaps = 0/115 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
TIGKR ASI VEN E+NR A R LLF+T G SG ILFEETLYQKS +GTP V++L+
Sbjct 35 TIGKRLASINVENVESNRRALRELLFTTPGALPCLSGVILFEETLYQKSSDGTPFVDMLK 94
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
G++PGIKVDKG + GT GE T GLDGL +RC+KYYEAGARFAKWRAVL+I
Sbjct 95 SAGVLPGIKVDKGTVELAGTNGETTTQGLDGLGDRCKKYYEAGARFAKWRAVLKI 149
> At4g26520
Length=358
Score = 157 bits (397), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 76/117 (64%), Positives = 88/117 (75%), Gaps = 0/117 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
TIGKRFA I VENTE+NR AYR LLF++ G SG ILFEETLYQK+ +G P V+LL
Sbjct 35 TIGKRFAGINVENTESNRQAYRELLFTSPGSYPCLSGVILFEETLYQKTSDGKPFVDLLM 94
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQIDA 117
+ G+IPGIKVDKGL + GT GE T GLD L RC++YYEAGARFAKWRA +I A
Sbjct 95 ENGVIPGIKVDKGLVDLAGTNGETTTQGLDSLGARCQQYYEAGARFAKWRAFFKIGA 151
> At4g26530
Length=358
Score = 156 bits (394), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 76/117 (64%), Positives = 89/117 (76%), Gaps = 0/117 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
TIGKRFASI VEN E+NR A R LLF++ G SG ILFEETLYQK+ +G P VELL
Sbjct 35 TIGKRFASINVENIESNRQALRELLFTSPGTFPCLSGVILFEETLYQKTTDGKPFVELLM 94
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQIDA 117
+ G+IPGIKVDKG+ + GT GE T GLD L RC++YY+AGARFAKWRAVL+I A
Sbjct 95 ENGVIPGIKVDKGVVDLAGTNGETTTQGLDSLGARCQEYYKAGARFAKWRAVLKIGA 151
> At2g21330
Length=393
Score = 149 bits (376), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 72/115 (62%), Positives = 85/115 (73%), Gaps = 0/115 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
T GKR ASIG+ENTEANR AYR LL S GLG+Y SGAILFEETLYQ + +G MV++L
Sbjct 86 TCGKRLASIGLENTEANRQAYRTLLVSAPGLGQYISGAILFEETLYQSTTDGKKMVDVLV 145
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+Q I+PGIKVDKGL + G+ E GLDGL+ R YY+ GARFAKWR V+ I
Sbjct 146 EQNIVPGIKVDKGLVPLVGSYDESWCQGLDGLASRTAAYYQQGARFAKWRTVVSI 200
> At3g52930
Length=358
Score = 149 bits (376), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 73/115 (63%), Positives = 85/115 (73%), Gaps = 0/115 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
TIGKR ASI VEN E NR R LLF+ G SG ILFEETLYQKS +G V++L+
Sbjct 35 TIGKRLASINVENVETNRRNLRELLFTAPGALPCLSGVILFEETLYQKSSDGKLFVDILK 94
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+ G++PGIKVDKG + GT GE T GLDGL +RC+KYYEAGARFAKWRAVL+I
Sbjct 95 EGGVLPGIKVDKGTVELAGTDGETTTQGLDGLGDRCKKYYEAGARFAKWRAVLKI 149
> At4g38970
Length=381
Score = 148 bits (374), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 75/128 (58%), Positives = 88/128 (68%), Gaps = 0/128 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
T GKR SIG+ENTEANR A+R LL S GLG+Y SGAILFEETLYQ + EG MV++L
Sbjct 78 TCGKRLDSIGLENTEANRQAFRTLLVSAPGLGQYVSGAILFEETLYQSTTEGKKMVDVLV 137
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQIDAAAA 120
+Q I+PGIKVDKGL + G+ E GLDGLS R YY+ GARFAKWR V+ I +
Sbjct 138 EQNIVPGIKVDKGLVPLVGSNNESWCQGLDGLSSRTAAYYQQGARFAKWRTVVSIPNGPS 197
Query 121 GGAAAAAA 128
A AA
Sbjct 198 ALAVKEAA 205
> At5g03690
Length=359
Score = 143 bits (360), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 74/115 (64%), Positives = 89/115 (77%), Gaps = 0/115 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
TIGKRF SI VEN E+NR A R LLF+T G +Y SG ILFEETLYQK+ G V++++
Sbjct 35 TIGKRFVSINVENVESNRRALRELLFTTPGALQYISGIILFEETLYQKTASGKLFVDVMK 94
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+ G++PGIKVDKG + GT GE T GLDGL +RC+KYYEAGARFAKWRAVL+I
Sbjct 95 EAGVLPGIKVDKGTVELAGTNGETTTTGLDGLGDRCKKYYEAGARFAKWRAVLKI 149
> At2g01140
Length=391
Score = 141 bits (356), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 69/128 (53%), Positives = 87/128 (67%), Gaps = 0/128 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
T GKR ASIG++NTE NR AYR LL +T GLG+Y SG+ILFEETLYQ + +G V+ L+
Sbjct 71 TCGKRLASIGLDNTEDNRQAYRQLLLTTPGLGDYISGSILFEETLYQSTKDGKTFVDCLR 130
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQIDAAAA 120
I+PGIKVDKGL + G+ E GLDGL+ R +YY+ GARFAKWR V+ + +
Sbjct 131 DANIVPGIKVDKGLSPLAGSNEESWCQGLDGLASRSAEYYKQGARFAKWRTVVSVPCGPS 190
Query 121 GGAAAAAA 128
A AA
Sbjct 191 ALAVKEAA 198
> CE16341
Length=365
Score = 137 bits (344), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 65/118 (55%), Positives = 83/118 (70%), Gaps = 1/118 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFST-KGLGEYCSGAILFEETLYQKSPEGTPMVELL 59
++ KR SIG+ENTE NR YR LLF+ L +Y SG I+F ET YQK+ +G P LL
Sbjct 39 SMDKRLNSIGLENTEENRRKYRQLLFTAGADLNKYISGVIMFHETFYQKTDDGKPFTALL 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQIDA 117
Q+QGIIPGIKVDKG+ + GT+GE T GLD L+ RC +Y + GA+FAKWR V +I +
Sbjct 99 QEQGIIPGIKVDKGVVPMAGTIGEGTTQGLDDLNARCAQYKKDGAQFAKWRCVHKISS 156
> 7301454
Length=361
Score = 135 bits (340), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 68/116 (58%), Positives = 82/116 (70%), Gaps = 1/116 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTK-GLGEYCSGAILFEETLYQKSPEGTPMVELL 59
T+GKR IGVENTE NR AYR LLFST L E SG ILF ETLYQK+ +GTP E+L
Sbjct 39 TMGKRLQDIGVENTEDNRRAYRQLLFSTDPKLAENISGVILFHETLYQKADDGTPFAEIL 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+++GII GIKVDKG+ + G+ E T GLD L+ RC +Y + G FAKWR VL+I
Sbjct 99 KKKGIILGIKVDKGVVPLFGSEDEVTTQGLDDLAARCAQYKKDGCDFAKWRCVLKI 154
> 7301455
Length=395
Score = 135 bits (339), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 68/116 (58%), Positives = 82/116 (70%), Gaps = 1/116 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTK-GLGEYCSGAILFEETLYQKSPEGTPMVELL 59
T+GKR IGVENTE NR AYR LLFST L E SG ILF ETLYQK+ +GTP E+L
Sbjct 39 TMGKRLQDIGVENTEDNRRAYRQLLFSTDPKLAENISGVILFHETLYQKADDGTPFAEIL 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+++GII GIKVDKG+ + G+ E T GLD L+ RC +Y + G FAKWR VL+I
Sbjct 99 KKKGIILGIKVDKGVVPLFGSEDEVTTQGLDDLAARCAQYKKDGCDFAKWRCVLKI 154
> Hs4885063
Length=364
Score = 130 bits (327), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 64/116 (55%), Positives = 77/116 (66%), Gaps = 1/116 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYC-SGAILFEETLYQKSPEGTPMVELL 59
++ KR + IGVENTE NR YR +LFS + C G I F ETLYQK G P V +
Sbjct 39 SMAKRLSQIGVENTEENRRLYRQVLFSADDRVKKCIGGVIFFHETLYQKDDNGVPFVRTI 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
Q +GI+ GIKVDKG+ + GT GE T GLDGLSERC +Y + GA FAKWR VL+I
Sbjct 99 QDKGIVVGIKVDKGVVPLAGTDGETTTQGLDGLSERCAQYKKDGADFAKWRCVLKI 154
> CE01225
Length=366
Score = 125 bits (313), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 58/115 (50%), Positives = 77/115 (66%), Gaps = 0/115 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
TIGKR +I +EN E NR YR LLF+T L ++ SG IL+EET +Q + +G +LL
Sbjct 43 TIGKRLDAINLENNETNRQKYRQLLFTTPNLNQHISGVILYEETFHQSTDKGEKFTDLLI 102
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+QGI+PGIK+D G+ + GT+GE T GLD L+ER + + G FAKWR VL I
Sbjct 103 KQGIVPGIKLDLGVVPLAGTIGEGTTQGLDKLAERAAAFKKGGCGFAKWRCVLNI 157
> Hs4557305
Length=364
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 62/116 (53%), Positives = 77/116 (66%), Gaps = 1/116 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYC-SGAILFEETLYQKSPEGTPMVELL 59
+I KR SIG ENTE NR YR LL + C G ILF ETLYQK+ +G P +++
Sbjct 39 SIAKRLQSIGTENTEENRRFYRQLLLTADDRVNPCIGGVILFHETLYQKADDGRPFPQVI 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+ +G + GIKVDKG+ + GT GE T GLDGLSERC +Y + GA FAKWR VL+I
Sbjct 99 KSKGGVVGIKVDKGVVPLAGTNGETTTQGLDGLSERCAQYKKDGADFAKWRCVLKI 154
> 7301463
Length=376
Score = 120 bits (300), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 57/116 (49%), Positives = 77/116 (66%), Gaps = 1/116 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTK-GLGEYCSGAILFEETLYQKSPEGTPMVELL 59
+GKRF IGVENTE NR YR +LF+T + E SG I + ETL+Q++ +G P VE L
Sbjct 39 VMGKRFQLIGVENTEENRRLYRQMLFTTDPKIAENISGVIFYHETLHQRTDDGLPFVEAL 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+++GI+ GIKVDK + G+ E T GLD L+ RC +Y + G FAKWR +L+I
Sbjct 99 RKKGILTGIKVDKHFSPLFGSEDEFTTQGLDDLANRCAQYKKEGCSFAKWRCILKI 154
> Hs14738249
Length=364
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/116 (48%), Positives = 73/116 (62%), Gaps = 1/116 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKG-LGEYCSGAILFEETLYQKSPEGTPMVELL 59
T+G R I VENTE NR +R +LFS + + G ILF ETLYQK +G +L
Sbjct 39 TMGNRLQRIKVENTEENRRQFREILFSVDSSINQSIGGVILFHETLYQKDSQGKLFRNIL 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+++GI+ GIK+D+G + GT E GLDGLSERC +Y + G F KWRAVL+I
Sbjct 99 KEKGIVVGIKLDQGGAPLAGTNKETTIQGLDGLSERCAQYKKDGVDFGKWRAVLRI 154
> Hs4557307
Length=364
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/116 (48%), Positives = 73/116 (62%), Gaps = 1/116 (0%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKG-LGEYCSGAILFEETLYQKSPEGTPMVELL 59
T+G R I VENTE NR +R +LFS + + G ILF ETLYQK +G +L
Sbjct 39 TMGNRLQRIKVENTEENRRQFREILFSVDSSINQSIGGVILFHETLYQKDSQGKLFRNIL 98
Query 60 QQQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVLQI 115
+++GI+ GIK+D+G + GT E GLDGLSERC +Y + G F KWRAVL+I
Sbjct 99 KEKGIVVGIKLDQGGAPLAGTNKETTIQGLDGLSERCAQYKKDGVDFGKWRAVLRI 154
> ECU01g0240
Length=338
Score = 101 bits (252), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 52/113 (46%), Positives = 72/113 (63%), Gaps = 2/113 (1%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYCSGAILFEETLYQKSPEGTPMVELLQ 60
T+G+RF +G+ NTE NR +R +LFSTKG+ Y G IL +ET Q S G P+ ELL+
Sbjct 33 TLGRRFEKLGITNTEENRRKFREILFSTKGIERYIGGVILNQETFEQTSGSGVPLTELLK 92
Query 61 QQGIIPGIKVDKGLETIPGTLGEQATMGLDGLSERCRKYYEAGARFAKWRAVL 113
++GI GIK+DKGL I E+ ++GL+ L RC+ A FAKWR++
Sbjct 93 KKGIEIGIKLDKGL--IDYKEKEKISVGLEDLDLRCKSSAFKDATFAKWRSLF 143
> Hs22045887
Length=258
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query 1 TIGKRFASIGVENTEANRAAYRGLLFSTKGLGEYC-SGAILFEETLYQKSPEGTPMVELL 59
+ K SIG ENTE NR YR L + C G ILF ETLYQK+ +G P +++
Sbjct 84 STAKWLQSIGTENTEENRCFYRQLWLTADNRVNPCIKGVILFHETLYQKADDGRPFPQVI 143
Query 60 QQQGIIPGIK 69
+ +G + IK
Sbjct 144 KSKGNVVSIK 153
> Hs7656985
Length=1669
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 65 IPGIKVDKGLETIPGTLGEQATMGLDGL 92
+PG+K GL IPGT GE+ ++G+ G+
Sbjct 738 LPGLKGLPGLPGIPGTPGEKGSIGVPGV 765
> CE28727
Length=1235
Score = 28.9 bits (63), Expect = 3.2, Method: Composition-based stats.
Identities = 32/104 (30%), Positives = 48/104 (46%), Gaps = 13/104 (12%)
Query 36 SGAILFEETLYQKSPEGTPMVELLQQ------QGIIPGIKVDKGLETIPGTLGEQATMG- 88
S + L T+Y++S T ++ +L + IIP K GL P L + G
Sbjct 643 SASFLTVSTMYRESL--TSLMTMLHTTHPHFIRCIIPNEKKTSGLIDAPLVLNQLTCNGV 700
Query 89 LDGLSERCRKYYEAGARFAKWR---AVLQIDAAAAGGAAAAAAA 129
L+G+ CRK + FA +R A+L D A+ AA A+ A
Sbjct 701 LEGI-RICRKGFPNRMMFADFRFRYAILAADEASDKDAAKASKA 743
> CE04334
Length=1758
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 52 GTPMVELLQQQGIIPGIKVDKGLETIPGTLGEQA 85
G P + + QG +PGI D+G++ PG GE
Sbjct 1155 GVPGIRGDKGQGGLPGIPGDRGMDGYPGQKGENG 1188
> CE04335
Length=1759
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 52 GTPMVELLQQQGIIPGIKVDKGLETIPGTLGEQA 85
G P + + QG +PGI D+G++ PG GE
Sbjct 1156 GVPGIRGDKGQGGLPGIPGDRGMDGYPGQKGENG 1189
> CE22614
Length=458
Score = 27.7 bits (60), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 20/48 (41%), Gaps = 8/48 (16%)
Query 66 PGIKVDKGLETIPGTLGEQATMGLDGLSERCRKY--------YEAGAR 105
PG + G++ +PG G T G G C KY YE G R
Sbjct 410 PGADGEPGMDGVPGNPGRDGTHGHGGEKGVCPKYCSADGGVFYEDGTR 457
Lambda K H
0.315 0.133 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1393086308
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40