bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1288_orf1
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
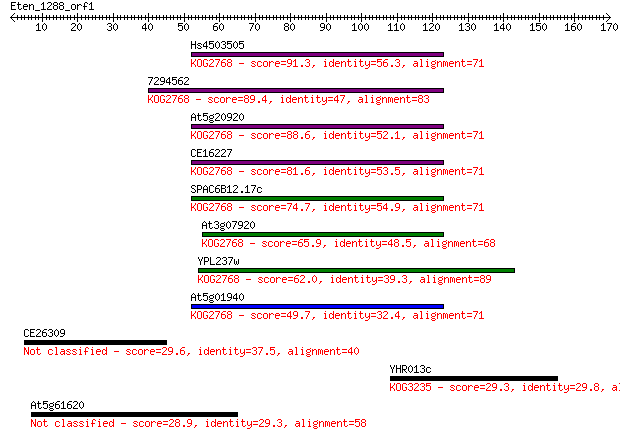
Hs4503505 91.3 8e-19
7294562 89.4 3e-18
At5g20920 88.6 5e-18
CE16227 81.6 7e-16
SPAC6B12.17c 74.7 9e-14
At3g07920 65.9 4e-11
YPL237w 62.0 5e-10
At5g01940 49.7 3e-06
CE26309 29.6 2.7
YHR013c 29.3 3.5
At5g61620 28.9 4.6
> Hs4503505
Length=333
Score = 91.3 bits (225), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 40/73 (54%), Positives = 57/73 (78%), Gaps = 2/73 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDL-AGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPS 109
Y+YEE+L R+ ++ NPD+ AG KR + +KPPQVVRVG+KK +++NF DIC ++HR
Sbjct 174 YTYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKKTSFVNFTDICKLLHRQP 233
Query 110 EHVLQFVLAELGT 122
+H+L F+LAELGT
Sbjct 234 KHLLAFLLAELGT 246
> 7294562
Length=312
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 39/85 (45%), Positives = 59/85 (69%), Gaps = 2/85 (2%)
Query 40 DGSGQLFVRGHVYSYEEMLKRIQELIVRNNPDLAGSKR--YTIKPPQVVRVGSKKVAWIN 97
D S F Y+Y+E+LKR+ E+I+ NPD+A ++ + ++PPQV+RVG+KK ++ N
Sbjct 141 DNSSTWFGSDRDYTYDELLKRVFEIILDKNPDMAAGRKPKFVMRPPQVLRVGTKKTSFAN 200
Query 98 FKDICGIMHRPSEHVLQFVLAELGT 122
F DI +HR +H+L F+LAELGT
Sbjct 201 FMDIAKTLHRLPKHLLDFLLAELGT 225
> At5g20920
Length=268
Score = 88.6 bits (218), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 37/72 (51%), Positives = 55/72 (76%), Gaps = 1/72 (1%)
Query 52 YSYEEMLKRIQELIVRNNPDLAGSKRYTI-KPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y Y+E+L R+ ++ NNP+LAG +R T+ +PPQV+R G+KK ++NF D+C MHR +
Sbjct 116 YIYDELLGRVFNILRENNPELAGDRRRTVMRPPQVLREGTKKTVFVNFMDLCKTMHRQPD 175
Query 111 HVLQFVLAELGT 122
HV+Q++LAELGT
Sbjct 176 HVMQYLLAELGT 187
> CE16227
Length=250
Score = 81.6 bits (200), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 51/72 (70%), Gaps = 1/72 (1%)
Query 52 YSYEEMLKRIQELIVRNNPDLAGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y+YEE L + +++ NPD AG K+ + IK P+V R GSKK A+ NF +IC +M R +
Sbjct 87 YTYEEALTLVYQVMKDKNPDFAGDKKKFAIKLPEVARAGSKKTAFSNFLEICRLMKRQDK 146
Query 111 HVLQFVLAELGT 122
HVLQF+LAELGT
Sbjct 147 HVLQFLLAELGT 158
> SPAC6B12.17c
Length=310
Score = 74.7 bits (182), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 39/72 (54%), Positives = 46/72 (63%), Gaps = 2/72 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDLAGSKR-YTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y Y E+L R L+ NNP+LAG KR YTI PP V R G KK + N DI MHR +
Sbjct 157 YYYPELLNRFFTLLRTNNPELAGEKRKYTIVPPSVHREG-KKTIFANISDISKRMHRSLD 215
Query 111 HVLQFVLAELGT 122
HV+QF+ AELGT
Sbjct 216 HVIQFLFAELGT 227
> At3g07920
Length=169
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 48/73 (65%), Gaps = 5/73 (6%)
Query 55 EEMLKRIQELIVRNNPDLAGSKRYTIK-PPQVVRV----GSKKVAWINFKDICGIMHRPS 109
+E+L+R+ ++ N+P+L G TI PPQV+R G+KK ++NF D C MHR
Sbjct 17 QEILRRVFNILRENSPELVGIWLLTIIWPPQVLREETAKGTKKTVFVNFMDYCKTMHRNP 76
Query 110 EHVLQFVLAELGT 122
+HV+ F+LAELGT
Sbjct 77 DHVMAFLLAELGT 89
> YPL237w
Length=285
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 51/92 (55%), Gaps = 15/92 (16%)
Query 54 YEEMLKRIQELIVRNNPDLAGSK---RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y E+L R ++ NNP+LAG + ++ I PP +R G KK + N +DI +HR E
Sbjct 131 YSELLSRFFNILRTNNPELAGDRSGPKFRIPPPVCLRDG-KKTIFSNIQDIAEKLHRSPE 189
Query 111 HVLQFVLAELGTESPQSNRPPKHLTSVKGEKR 142
H++Q++ AELGT SV G+KR
Sbjct 190 HLIQYLFAELGTSG-----------SVDGQKR 210
> At5g01940
Length=231
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 52 YSYEEMLKRIQELIVRNNPDLAGSK-RYTIKPPQVVRVGSKKVAWINFKDICGIMHRPSE 110
Y Y+E+L + + + + +++ + R + PPQ++ G+ V +NF D+C MHR +
Sbjct 73 YGYKELLSMVFDRLREEDVEVSTERPRTVMMPPQLLAEGTITVC-LNFADLCRTMHRKPD 131
Query 111 HVLQFVLAELGT 122
HV++F+LA++ T
Sbjct 132 HVMKFLLAQMET 143
> CE26309
Length=526
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 5 EHRAAQAFDFGERRKKKKEKKPEDQA-AEAASEAFVDGSGQ 44
+ +++Q R + KK K+PED+A ++ +E VDG+GQ
Sbjct 207 KKQSSQEVGGASRSEDKKTKEPEDEAESDTVTEKKVDGAGQ 247
> YHR013c
Length=238
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Query 108 PSEHVLQFVLAELGTESPQSNRPPK-HLTSVKGEKRKRKRSQKTRLLR 154
P E ++ +VL ++ + Q N PP H+TS+ + R+ L+R
Sbjct 88 PGEKLVGYVLVKMNDDPDQQNEPPNGHITSLSVMRTYRRMGIAENLMR 135
> At5g61620
Length=317
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query 7 RAAQAFDFGERRKKKKEKKPEDQAAEAASEAFVDGSGQLFVRGHVYSYEEMLKRIQEL 64
R A FD +K+KE+ +D + + + + G Q V+GH + E+ R Q L
Sbjct 165 RRASLFDISLEDQKEKERNSQDASTKTPPKQPITGIQQPVVQGHTQT--EISNRFQNL 220
Lambda K H
0.317 0.132 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2454498488
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40