bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1300_orf2
Length=201
Score E
Sequences producing significant alignments: (Bits) Value
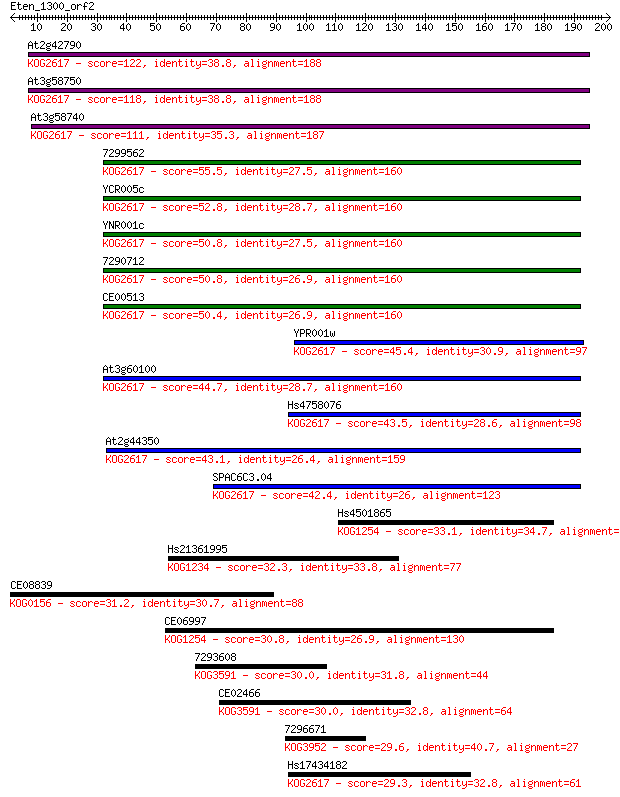
At2g42790 122 7e-28
At3g58750 118 8e-27
At3g58740 111 1e-24
7299562 55.5 8e-08
YCR005c 52.8 5e-07
YNR001c 50.8 2e-06
7290712 50.8 2e-06
CE00513 50.4 2e-06
YPR001w 45.4 8e-05
At3g60100 44.7 1e-04
Hs4758076 43.5 3e-04
At2g44350 43.1 3e-04
SPAC6C3.04 42.4 7e-04
Hs4501865 33.1 0.34
Hs21361995 32.3 0.70
CE08839 31.2 1.3
CE06997 30.8 1.9
7293608 30.0 2.9
CE02466 30.0 3.2
7296671 29.6 4.4
Hs17434182 29.3 5.9
> At2g42790
Length=509
Score = 122 bits (305), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 73/200 (36%), Positives = 110/200 (55%), Gaps = 14/200 (7%)
Query 7 VLRTACSVLGSLEPERNPTQQQVDI----------ALRLIGCLPGALSYWFLYTNSGKEI 56
VL +A S L P+ NP + DI +R+IG P + +L +G+
Sbjct 181 VLVSAMSALSIFHPDANPALRGQDIYDSKQVRDKQIIRIIGKAPTIAAAAYLRM-AGRPP 239
Query 57 SFQSDPEDSVATNFLKLLH--GDPSFKPHPVVVKTLDTSLILYAEHDFNASTFACRVTAG 114
S A NFL +L G+ S+KP+P + + LD IL+AEH+ N ST A R A
Sbjct 240 VLPSG-NLPYADNFLYMLDSLGNRSYKPNPRLARVLDILFILHAEHEMNCSTAAARHLAS 298
Query 115 TRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLTSIQKAQRRLEEAFSRRELIMGFGH 174
+ D+Y+ + +G L GPLHGGANEA LKML + +++ +E +R+ + GFGH
Sbjct 299 SGVDVYTAVAGAVGALYGPLHGGANEAVLKMLSEIGTVENIPEFIEGVKNRKRKMSGFGH 358
Query 175 RMYKSGDPRAPLMRGLAEAL 194
R+YK+ DPRA +++ LA+ +
Sbjct 359 RVYKNYDPRAKVIKNLADEV 378
> At3g58750
Length=514
Score = 118 bits (296), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 73/200 (36%), Positives = 109/200 (54%), Gaps = 14/200 (7%)
Query 7 VLRTACSVLGSLEPERNPTQQQVDI----------ALRLIGCLPGALSYWFLYTNSGKEI 56
VL +A S L P+ NP DI +R++G P + +L T +G+
Sbjct 186 VLVSAMSALSIFHPDANPALSGQDIYKSKQVRDKQIVRILGKAPTIAAAAYLRT-AGRPP 244
Query 57 SFQSDPEDSVATNFLKLLH--GDPSFKPHPVVVKTLDTSLILYAEHDFNASTFACRVTAG 114
S S + NFL +L G+ S+KP+P + + LD IL+AEH+ N ST A R A
Sbjct 245 VLPS-ANLSYSENFLYMLDSMGNRSYKPNPRLARVLDILFILHAEHEMNCSTAAARHLAS 303
Query 115 TRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLTSIQKAQRRLEEAFSRRELIMGFGH 174
+ D+Y+ +G L GPLHGGANEA LKML + + + +E +R+ + GFGH
Sbjct 304 SGVDVYTACAGAVGALYGPLHGGANEAVLKMLAEIGTAENIPDFIEGVKNRKRKMSGFGH 363
Query 175 RMYKSGDPRAPLMRGLAEAL 194
R+YK+ DPRA +++ LA+ +
Sbjct 364 RVYKNYDPRAKVIKKLADEV 383
> At3g58740
Length=480
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 66/199 (33%), Positives = 107/199 (53%), Gaps = 14/199 (7%)
Query 8 LRTACSVLGSLEPERNPT----------QQQVDIALRLIGCLPGALSYWFLYTNSGKEIS 57
L TA S L P+ NP+ Q + +R++G P + +L +GK
Sbjct 184 LVTAMSALSIFYPDANPSLMGLGVYKSKQVRDKQIVRVLGQAPTIAAAAYL-RKAGKP-P 241
Query 58 FQSDPEDSVATNFLKLLH--GDPSFKPHPVVVKTLDTSLILYAEHDFNASTFACRVTAGT 115
Q S + NFL ++ GD S+KP+P + + LD IL AEH+ N ST A R + +
Sbjct 242 VQPLSNLSYSENFLYMVESMGDRSYKPNPRLARVLDILFILQAEHEMNCSTAAARHLSSS 301
Query 116 RSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLTSIQKAQRRLEEAFSRRELIMGFGHR 175
D+Y+ + G+G + GPLHGGA EA + ML + +++ +E +++ + GFGHR
Sbjct 302 GGDVYTAVSGGVGAIYGPLHGGAVEATINMLSEIGTVENIPEFIESVKNKKRRLSGFGHR 361
Query 176 MYKSGDPRAPLMRGLAEAL 194
+YK+ DPR +++ LA+ +
Sbjct 362 IYKNYDPRGKVVKKLADEV 380
> 7299562
Length=407
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 44/171 (25%), Positives = 79/171 (46%), Gaps = 19/171 (11%)
Query 32 ALRLIGCLPGALSYWFLYTNSGKEISFQSDPEDSVATNFLKLLHGDPSFKPHPVVVKTLD 91
++ LI LP + + E S + + E+ + NF ++L P V +
Sbjct 132 SMNLIAMLPTVAAAIYSNVFRDGEGSREVNYEEDWSGNFCRMLG-----LPEKDFVDLMR 186
Query 92 TSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLT 150
+IL+A+H+ N S AC + SD + + + L GPLHG AN+ ++L+ LT
Sbjct 187 LYMILHADHESGNVSAHACHLVGTALSDPFLSFSASMCGLAGPLHGLANQ---EVLVWLT 243
Query 151 SIQKA----------QRRLEEAFSRRELIMGFGHRMYKSGDPRAPLMRGLA 191
++KA ++ +++ ++I G+GH + + DPR L A
Sbjct 244 KLRKAIGDDPSDEELKKFIDDTLKGGQVIPGYGHAVLRDTDPRFVLQNEFA 294
> YCR005c
Length=460
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 46/170 (27%), Positives = 73/170 (42%), Gaps = 18/170 (10%)
Query 32 ALRLIGCLPGALSYWFLYTNSGKEISF-QSDPEDSVATNFLKLL-HGDPSFKPHPVVVKT 89
+L L+G LP + +Y N K+ + DP A N + L+ D F V
Sbjct 195 SLDLLGKLPVIAAK--IYRNVFKDGKMGEVDPNADYAKNLVNLIGSKDEDF------VDL 246
Query 90 LDTSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIP 148
+ L ++++H+ N S + S Y + SG+ L GPLHG AN+ L+ L
Sbjct 247 MRLYLTIHSDHEGGNVSAHTSHLVGSALSSPYLSLASGLNGLAGPLHGRANQEVLEWLFA 306
Query 149 LT-------SIQKAQRRLEEAFSRRELIMGFGHRMYKSGDPRAPLMRGLA 191
L S ++ L + + +I G+GH + + DPR R A
Sbjct 307 LKEEVNDDYSKDTIEKYLWDTLNSGRVIPGYGHAVLRKTDPRYMAQRKFA 356
> YNR001c
Length=479
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 44/170 (25%), Positives = 74/170 (43%), Gaps = 18/170 (10%)
Query 32 ALRLIGCLPGALSYWFLYTNSGKEISFQS-DPEDSVATNFLKLL-HGDPSFKPHPVVVKT 89
+L L+G LP S +Y N K+ S DP N +LL + + F +
Sbjct 214 SLDLLGKLPVIASK--IYRNVFKDGKITSTDPNADYGKNLAQLLGYENKDF------IDL 265
Query 90 LDTSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIP 148
+ L ++++H+ N S + S Y + +G+ L GPLHG AN+ L+ L
Sbjct 266 MRLYLTIHSDHEGGNVSAHTTHLVGSALSSPYLSLAAGLNGLAGPLHGRANQEVLEWLFK 325
Query 149 LT-------SIQKAQRRLEEAFSRRELIMGFGHRMYKSGDPRAPLMRGLA 191
L S + ++ L + + ++ G+GH + + DPR R A
Sbjct 326 LREEVKGDYSKETIEKYLWDTLNAGRVVPGYGHAVLRKTDPRYTAQREFA 375
> 7290712
Length=464
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/171 (25%), Positives = 78/171 (45%), Gaps = 19/171 (11%)
Query 32 ALRLIGCLPGALSYWFLYTNSGKEISFQSDPEDSVATNFLKLLHGDPSFKPHPVVVKTLD 91
++ LI LP + + T G + S D + NF+K+L D + + +
Sbjct 203 SMDLIAKLPVVAATIYCNTYRGGKGSRSIDSSLDWSANFVKMLGYD-----NAPFTELMR 257
Query 92 TSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLT 150
L ++++H+ N S + SD Y +G+ L GPLHG AN+ ++L+ L
Sbjct 258 LYLTIHSDHEGGNVSAHTVHLVGSALSDPYLSFAAGLNGLAGPLHGLANQ---EVLVWLR 314
Query 151 SIQK------AQRRLEEAFSRR----ELIMGFGHRMYKSGDPRAPLMRGLA 191
+QK ++ +L+E + +++ G+GH + + DPR R A
Sbjct 315 KLQKEAGNNPSEEQLKEYIWKTLKSGQVVPGYGHAVLRKTDPRYTCQREFA 365
> CE00513
Length=468
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/170 (25%), Positives = 76/170 (44%), Gaps = 17/170 (10%)
Query 32 ALRLIGCLP--GALSYWFLYTNSGKEISFQSDPEDSVATNFLKLLHGDPSFKPHPVVVKT 89
++ L+ LP A+ Y LY + G +S DP+ + NF +L D P+ +
Sbjct 204 SMDLLAKLPTVAAIIYRNLYRD-GSAVSV-IDPKKDWSANFSSMLGYD-----DPLFAEL 256
Query 90 LDTSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIP 148
+ L+++++H+ N S + SD Y + + L GPLHG AN+ L L
Sbjct 257 MRLYLVIHSDHEGGNVSAHTSHLVGSALSDPYLSFSAAMAGLAGPLHGLANQEVLVFLNK 316
Query 149 LT---SIQKAQRRLEEAFSRR----ELIMGFGHRMYKSGDPRAPLMRGLA 191
+ + +L+E + +++ G+GH + + DPR R A
Sbjct 317 IVGEIGFNYTEEQLKEWVWKHLKSGQVVPGYGHAVLRKTDPRYECQREFA 366
> YPR001w
Length=486
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 48/105 (45%), Gaps = 8/105 (7%)
Query 96 LYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLTSI-- 152
++ +H+ N S + SD Y SGI L GPLHG A + ++ LI + S
Sbjct 275 IHVDHEGGNVSAHTTHLVGSALSDPYLSYSSGIMGLAGPLHGLAAQEVVRFLIEMNSNIS 334
Query 153 -----QKAQRRLEEAFSRRELIMGFGHRMYKSGDPRAPLMRGLAE 192
Q+ + L + + +I G+GH + + DPR M A+
Sbjct 335 SIAREQEIKDYLWKILNSNRVIPGYGHAVLRKPDPRFTAMLEFAQ 379
> At3g60100
Length=433
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 46/169 (27%), Positives = 73/169 (43%), Gaps = 16/169 (9%)
Query 32 ALRLIGCLPGALSYWFLYTNSGKEISFQSDPEDSVATNFLKLLHGDPSFKPHPVVVKTLD 91
AL LI +P SY + I D D A NF +L D P + + +
Sbjct 171 ALNLIARVPVVASYVYRRMYKDGSIIPLDDSLDYGA-NFSHMLGFD-----SPQMKELMR 224
Query 92 TSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLT 150
+ ++++H+ N S A + SD Y + + L GPLHG AN+ L + I L
Sbjct 225 LYVTIHSDHEGGNVSAHAGHLVGSALSDPYLSFAAALNGLAGPLHGLANQEVL-LWIKLV 283
Query 151 ------SIQKAQRR--LEEAFSRRELIMGFGHRMYKSGDPRAPLMRGLA 191
SI K Q + + + + +++ G+GH + + DPR R A
Sbjct 284 VEECGESISKEQLKDYVWKTLNSGKVVPGYGHGVLRKTDPRYICQREFA 332
> Hs4758076
Length=466
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 46/106 (43%), Gaps = 8/106 (7%)
Query 94 LILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPL--- 149
L ++++H+ N S + SD Y + + L GPLHG AN+ L L L
Sbjct 259 LTIHSDHEGGNVSAHTSHLVGSALSDPYLSFAAAMNGLAGPLHGLANQEVLVWLTQLQKE 318
Query 150 ----TSIQKAQRRLEEAFSRRELIMGFGHRMYKSGDPRAPLMRGLA 191
S +K + + + ++ G+GH + + DPR R A
Sbjct 319 VGKDVSDEKLRDYIWNTLNSGRVVPGYGHAVLRKTDPRYTCQREFA 364
> At2g44350
Length=474
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 71/169 (42%), Gaps = 18/169 (10%)
Query 33 LRLIGCLPGALSYWF--LYTNSGKEISFQSDPEDSVATNFLKLLHGDPSFKPHPVVVKTL 90
L LI +P +Y + +Y N S SD NF +L D V + +
Sbjct 211 LNLIARVPVVAAYVYRRMYKNGD---SIPSDKSLDYGANFSHMLGFDDE-----KVKELM 262
Query 91 DTSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPL 149
+ ++++H+ N S + SD Y + + L GPLHG AN+ L + +
Sbjct 263 RLYITIHSDHEGGNVSAHTGHLVGSALSDPYLSFAAALNGLAGPLHGLANQEVLLWIKSV 322
Query 150 T-----SIQKAQRR--LEEAFSRRELIMGFGHRMYKSGDPRAPLMRGLA 191
I K Q + + + + ++I G+GH + ++ DPR R A
Sbjct 323 VEECGEDISKEQLKEYVWKTLNSGKVIPGYGHGVLRNTDPRYVCQREFA 371
> SPAC6C3.04
Length=473
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 32/131 (24%), Positives = 56/131 (42%), Gaps = 12/131 (9%)
Query 69 NFLKLLHGDPSFKPHPVVVKTLDTSLILYAEHDF-NASTFACRVTAGTRSDIYSCICSGI 127
NF +L F + V+ + L ++A+H+ N S + S + + + +
Sbjct 247 NFANVL----GFANNEEFVELMRLYLTIHADHEGGNVSAHTGHLVGSALSSPFLSMAASL 302
Query 128 GTLKGPLHGGANEAALKMLIPLT-------SIQKAQRRLEEAFSRRELIMGFGHRMYKSG 180
L GPLHG AN+ L LI + S + + L + + ++ G+GH + +
Sbjct 303 NGLAGPLHGLANQEVLNFLITMKKEIGDDLSEETIKSYLWKLLNSGRVVPGYGHAVLRKT 362
Query 181 DPRAPLMRGLA 191
DPR R A
Sbjct 363 DPRYTAQREFA 373
> Hs4501865
Length=1105
Score = 33.1 bits (74), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 40/80 (50%), Gaps = 16/80 (20%)
Query 111 VTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKM--------LIPLTSIQKAQRRLEEA 162
+ A T ++ S + SG+ T+ G GGA +AA KM +IP+ + K ++
Sbjct 916 ICARTAVELVSSLTSGLLTI-GDRFGGALDAAAKMFSKAFDSGIIPMEFVNKMKKE---- 970
Query 163 FSRRELIMGFGHRMYKSGDP 182
+LIMG GHR+ +P
Sbjct 971 ---GKLIMGIGHRVKSINNP 987
> Hs21361995
Length=386
Score = 32.3 bits (72), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 40/79 (50%), Gaps = 6/79 (7%)
Query 54 KEISFQSD--PEDSVATNFLKLLHGDPSFKPHPVVVKTLDTSLILYAEHDFNASTFACRV 111
+E++++ D E + A NF +LL DP F+ P VVK L T+ +L E C+
Sbjct 121 QELAWECDYRREAACAQNFRQLLANDPFFRV-PAVVKELCTTRVLGMELAGGVPLDQCQ- 178
Query 112 TAGTRSDIYSCICSGIGTL 130
G D+ + IC + TL
Sbjct 179 --GLSQDLRNQICFQLLTL 195
> CE08839
Length=509
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 42/91 (46%), Gaps = 6/91 (6%)
Query 1 QKLYKDVLRTACS---VLGSLEPERNPTQQQVDIALRLIGCLPGALSYWFLYTNSGKEIS 57
QK+Y+++ R S + + P+ N V+ + RL LP LS TN+ EI+
Sbjct 340 QKVYEELEREIGSDRIITTTDRPKLNYINATVNESQRLANLLPMNLS---RSTNADVEIA 396
Query 58 FQSDPEDSVATNFLKLLHGDPSFKPHPVVVK 88
P+D+V T + + DP P P K
Sbjct 397 GYRIPKDTVITPQISSVMYDPEIFPEPYEFK 427
> CE06997
Length=1106
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 55/140 (39%), Gaps = 15/140 (10%)
Query 53 GKEISFQSDP------EDSVATNFLKLLHGDPSFKPHPVVVKTLDTSLILYAEHDFNAS- 105
G+E+++ P D L LL PH K ++ L+L A+H S
Sbjct 849 GEELNYAGVPITKVLESDMGIGGVLGLLWFQKRLPPH--ANKFIEICLMLTADHGPAVSG 906
Query 106 ---TFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLTSIQKAQRRLEEA 162
T C A D+ S + SG+ T+ G + AA + A + + E
Sbjct 907 AHNTIVC---ARAGKDLISSLTSGLLTIGDRFGGALDGAARQFSEAFDQGWSANQFVSEM 963
Query 163 FSRRELIMGFGHRMYKSGDP 182
+ + IMG GHR+ +P
Sbjct 964 RKKGKHIMGIGHRVKSINNP 983
> 7293608
Length=183
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 63 EDSVATNFLKLLHGDPSFKPHPVVVKTLDTSLILYAEHDFNAST 106
+D LKL + P +VVKT+D L+++A+H+ + T
Sbjct 87 QDEGDNKVLKLRFDVSQYAPEEIVVKTVDQKLLVHAKHEEKSDT 130
> CE02466
Length=219
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 35/74 (47%), Gaps = 10/74 (13%)
Query 71 LKLLHGDPSFKPHPVVVKTLDTSLILYAEHD--------FNASTFACRVTAGTRSDIYSC 122
L+L ++KP V VKT+D L+++A+H+ F + GT + S
Sbjct 139 LRLRFDVANYKPEEVTVKTIDNRLLVHAKHEEKTPQRTVFREYNQEFLLPRGTNPEQISS 198
Query 123 ICS--GIGTLKGPL 134
S G+ T++ PL
Sbjct 199 TLSTDGVLTVEAPL 212
> 7296671
Length=411
Score = 29.6 bits (65), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 93 SLILYAEHDFNASTFACRVTAGTRSDI 119
+ + Y EHDF A+ F+C AGT+ +
Sbjct 263 AFLYYLEHDFAATPFSCGQLAGTKKAV 289
> Hs17434182
Length=264
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Query 94 LILYAEHDF-NASTFACRVTAGTRSDIYSCICSGIGTLKGPLHGGANEAALKMLIPLTSI 152
L ++++H+ N S A + T SD + + L PLHG AN+ ++L+ LT +
Sbjct 174 LTIHSDHEHSNISAHARHLAGSTLSDPSLPFAAAMNGLAVPLHGLANQ---EVLVWLTQL 230
Query 153 QK 154
QK
Sbjct 231 QK 232
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3515233148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40