bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1304_orf3
Length=268
Score E
Sequences producing significant alignments: (Bits) Value
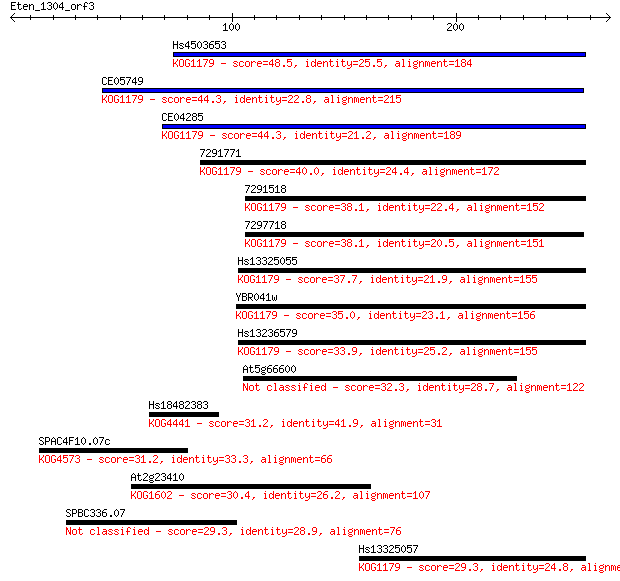
Hs4503653 48.5 1e-05
CE05749 44.3 3e-04
CE04285 44.3 3e-04
7291771 40.0 0.005
7291518 38.1 0.018
7297718 38.1 0.019
Hs13325055 37.7 0.026
YBR041w 35.0 0.15
Hs13236579 33.9 0.39
At5g66600 32.3 1.0
Hs18482383 31.2 2.2
SPAC4F10.07c 31.2 2.7
At2g23410 30.4 4.4
SPBC336.07 29.3 8.6
Hs13325057 29.3 9.2
> Hs4503653
Length=620
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 47/201 (23%), Positives = 80/201 (39%), Gaps = 33/201 (16%)
Query 74 LHHLSARSKTDE-----QKRDHMHERECPD-------MALPCCCLYTADLEGHLRATWVS 121
++++S S TD K D + P+ + P +YT+ G +A ++
Sbjct 179 IYYVSRTSNTDGIDSFLDKVDEVSTEPIPESWRSEVTFSTPALYIYTSGTTGLPKAAMIT 238
Query 122 NSRF-LSAGLCWVHAVDLTESDRLLLGVQLCHE----VGLHALAAAISCRGALLLRSRCS 176
+ R GL +V L D + + + H +G+H I L LR++ S
Sbjct 239 HQRIWYGTGLTFVSG--LKADDVIYITLPFYHSAALLIGIHG---CIVAGATLALRTKFS 293
Query 177 LLSLWPDVKALDATIIFHTGMLWSRLLKAHERFNAGDETQRLGSWVGHPQLRASIGTGLP 236
W D + + T+I + G L L + ++ N D RL ++G GL
Sbjct 294 ASQFWDDCRKYNVTVIQYIGELLRYLCNSPQKPNDRDHKVRL-----------ALGNGLR 342
Query 237 GELWPVVKSVFNIPRILEFYS 257
G++W F I EFY+
Sbjct 343 GDVWRQFVKRFGDICIYEFYA 363
> CE05749
Length=650
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 49/216 (22%), Positives = 89/216 (41%), Gaps = 24/216 (11%)
Query 42 DEVAYHKQHVEQHHDKEHKCKPSGFPLLLKQRLHHLSARSKTDEQKRDHMHERECPDMAL 101
D+ + + +E + E K K SGF L K+ L A+ T+ + D + D
Sbjct 193 DQKLFDVEGIEVYSVGEPK-KNSGFKNLKKK----LDAQITTEPKTLDIV------DFKS 241
Query 102 PCCCLYTADLEGHLRATWVSNSRFLSAGLCWVHAVDLTESDRLLLGVQLCHE-VGLHALA 160
C +YT+ G +A + + R+ S + + + SDR+ + + + H G+ +
Sbjct 242 ILCFIYTSGTTGMPKAAVMKHFRYYSIAVGAAKSFGIRPSDRMYVSMPIYHTAAGILGVG 301
Query 161 AAISCRGALLLRSRCSLLSLWPDVKALDATIIFHTGMLWSRLLKAHERFNAGDETQRLGS 220
A+ + ++R + S + W D D T+ + G + LL Q +
Sbjct 302 QALLGGSSCVIRKKFSASNFWRDCVKYDCTVSQYIGEICRYLL-----------AQPVVE 350
Query 221 WVGHPQLRASIGTGLPGELWPVVKSVFNIPRILEFY 256
++R +G GL E+W F + RI E Y
Sbjct 351 EESRHRMRLLVGNGLRAEIWQPFVDRFRV-RIGELY 385
> CE04285
Length=655
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/201 (19%), Positives = 79/201 (39%), Gaps = 23/201 (11%)
Query 69 LLKQRLHHLSARSKTDEQKRD-----HMHERECP------DMALPCCCLYTADLEGHLRA 117
L+ +H A ++ D + R H+ + P + C +YT+ G+ +
Sbjct 201 LISDEIHVFLAGTQVDGRHRSLQQDLHLFSEDEPPVIDGLNFRSVLCYIYTSGTTGNPKP 260
Query 118 TWVSNSRFLSAGLCWVHAVDLTESDRLLLGVQLCHEV-GLHALAAAISCRGALLLRSRCS 176
+ + R+ + A + +SD + + + + H G+ + + I+ ++R + S
Sbjct 261 AVIKHFRYFWIAMGAGKAFGINKSDVVYITMPMYHSAAGIMGIGSLIAFGSTAVIRKKFS 320
Query 177 LLSLWPDVKALDATIIFHTGMLWSRLLKAHERFNAGDETQRLGSWVGHPQLRASIGTGLP 236
+ W D + T + G + LL A+ +E Q +R G GL
Sbjct 321 ASNFWKDCVKYNVTATQYIGEICRYLLAANP---CPEEKQH--------NVRLMWGNGLR 369
Query 237 GELWPVVKSVFNIPRILEFYS 257
G++W F I +I E Y
Sbjct 370 GQIWKEFVGRFGIKKIGELYG 390
> 7291771
Length=661
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 42/174 (24%), Positives = 69/174 (39%), Gaps = 13/174 (7%)
Query 86 QKRDHMHERECPDMALP-CCCLYTADLEGHLRATWVSNSRFLSAGLCWVHAVDLTESDRL 144
QK+ + CP A +YT+ G +A ++N RFL + + ++ D +
Sbjct 242 QKKLELPSAVCPGEARSKLLYVYTSGTTGLPKAAVITNLRFLFMSAGSYYMLKMSSDDVV 301
Query 145 LLGVQLCHEVG-LHALAAAISCRGALLLRSRCSLLSLWPDVKALDATIIFHTGMLWSRLL 203
+ L H G + + AI ++LR + S + W D + T+ + G L LL
Sbjct 302 YDPLPLYHTAGGIVGVGNAILNGSTVVLRKKFSARNFWLDCDRHNCTVAQYIGELCRYLL 361
Query 204 KAHERFNAGDETQRLGSWVGHPQLRASIGTGLPGELWPVVKSVFNIPRILEFYS 257
+ D+ + LR G GL ++W F IP I E Y
Sbjct 362 ATSY---SPDQQKH--------NLRLMYGNGLRPQIWSQFVRRFGIPHIGEIYG 404
> 7291518
Length=664
Score = 38.1 bits (87), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 34/159 (21%), Positives = 67/159 (42%), Gaps = 24/159 (15%)
Query 106 LYTADLEGHLRATWVSNSR--FLSAGLCWVHAVDLTESDRLLLGVQLCHEVG-LHALAAA 162
+YT+ G +A +++SR F++AG+ + + + D + L H G + ++ A
Sbjct 273 IYTSGTTGLPKAAVITHSRYFFIAAGIHYT--LGFKDQDVFYTPLPLYHTAGGVMSMGQA 330
Query 163 ISCRGALLLRSRCSLLSLWPDVKALDATIIFHTGMLWSRLLKA----HERFNAGDETQRL 218
+ +++R + S + D T+ + G + +L H+R
Sbjct 331 LLFGSTVVIRKKFSASGYFSDCARFQCTVGQYIGEMARYILATPSAPHDR---------- 380
Query 219 GSWVGHPQLRASIGTGLPGELWPVVKSVFNIPRILEFYS 257
+ Q+R G GL ++WP F I ++ EFY
Sbjct 381 -----NHQVRMVFGNGLRPQIWPQFVERFGIRKVGEFYG 414
> 7297718
Length=690
Score = 38.1 bits (87), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 31/152 (20%), Positives = 59/152 (38%), Gaps = 24/152 (15%)
Query 106 LYTADLEGHLRATWVSNSRFLSAGLCWVHAVDLTESDRLLLGVQLCHEVG-LHALAAAIS 164
+YT+ G +A +S+SR+L + + E D + L H G + + ++
Sbjct 304 IYTSGTTGLPKAAVISHSRYLFIAAGIHYTMGFQEEDIFYTPLPLYHTAGGIMCMGQSVL 363
Query 165 CRGALLLRSRCSLLSLWPDVKALDATIIFHTGMLWSRLLKAHERFNAGDETQRLGSWVGH 224
+ +R + S + + D +AT+ T + +
Sbjct 364 FGSTVSIRKKFSASNYFADCAKYNATV-----------------------TTKPSEYDQK 400
Query 225 PQLRASIGTGLPGELWPVVKSVFNIPRILEFY 256
++R G GL ++WP FNI ++ EFY
Sbjct 401 HRVRLVFGNGLRPQIWPQFVQRFNIAKVGEFY 432
> Hs13325055
Length=619
Score = 37.7 bits (86), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 34/157 (21%), Positives = 62/157 (39%), Gaps = 15/157 (9%)
Query 103 CCCLYTADLEGHLRATWVSNSRFL-SAGLCWVHAVDLTESDRLLLGVQLCHE-VGLHALA 160
C ++T+ G +A +S + L + + W A T D + + + L H + ++
Sbjct 219 CLYIFTSGTTGLPKAAVISQLQVLRGSAVLW--AFGCTAHDIVYITLPLYHSSAAILGIS 276
Query 161 AAISCRGALLLRSRCSLLSLWPDVKALDATIIFHTGMLWSRLLKAHERFNAGDETQRLGS 220
+ +L+ + S W D K D T+ + G L L K +R D RL
Sbjct 277 GCVELGATCVLKKKFSASQFWSDCKKYDVTVFQYIGELCRYLCKQSKREGEKDHKVRL-- 334
Query 221 WVGHPQLRASIGTGLPGELWPVVKSVFNIPRILEFYS 257
+IG G+ ++W F ++ E Y+
Sbjct 335 ---------AIGNGIRSDVWREFLDRFGNIKVCELYA 362
> YBR041w
Length=623
Score = 35.0 bits (79), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 36/157 (22%), Positives = 63/157 (40%), Gaps = 12/157 (7%)
Query 102 PCCCLYTADLEGHLRATWVSNSRFLSAGLCWVHAVDLTESDRLLLGVQLCHEVG-LHALA 160
P +YT+ G ++ +S + + H + +T + + L H L
Sbjct 251 PSMLIYTSGTTGLPKSAIMSWRKSSVGCQVFGHVLHMTNESTVFTAMPLFHSTAALLGAC 310
Query 161 AAISCRGALLLRSRCSLLSLWPDVKALDATIIFHTGMLWSRLLKAHERFNAGDETQRLGS 220
A +S G L L + S + W V AT I + G + LL H + ++ ++
Sbjct 311 AILSHGGCLALSHKFSASTFWKQVYLTGATHIQYVGEVCRYLL--HTPISKYEKMHKV-- 366
Query 221 WVGHPQLRASIGTGLPGELWPVVKSVFNIPRILEFYS 257
+ + G GL ++W + FNI I EFY+
Sbjct 367 -------KVAYGNGLRPDIWQDFRKRFNIEVIGEFYA 396
> Hs13236579
Length=730
Score = 33.9 bits (76), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 39/162 (24%), Positives = 69/162 (42%), Gaps = 25/162 (15%)
Query 103 CCCLYTADLEGHLRATWVSNSRFLSAG----LCWVHAVDLTESDRLLLGVQLCHEVGLHA 158
C ++T+ G +A +S+ + L LC VH + D + L + L H G +
Sbjct 330 CLYIFTSGTTGLPKAARISHLKILQCQGFYQLCGVH-----QEDVIYLALPLYHMSG--S 382
Query 159 LAAAISCRG---ALLLRSRCSLLSLWPDVKALDATIIFHTGMLWSRLLKAHERFNAGDET 215
L + C G ++L+S+ S W D + T+ + G L L+ +
Sbjct 383 LLGIVGCMGIGATVVLKSKFSAGQFWEDCQQHRVTVFQYIGELCRYLVN-----QPPSKA 437
Query 216 QRLGSWVGHPQLRASIGTGLPGELWPVVKSVFNIPRILEFYS 257
+R GH ++R ++G+GL + W F ++LE Y
Sbjct 438 ER-----GH-KVRLAVGSGLRPDTWERFVRRFGPLQVLETYG 473
> At5g66600
Length=614
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 35/131 (26%), Positives = 59/131 (45%), Gaps = 12/131 (9%)
Query 105 CLYTADLEGHLRATWVSNSRFLSAGLCWVHAVDLTESDRLLLGVQLCHEVGLHA-LAAAI 163
C+Y + + N + L + L V L ++L + + + + +HA LA I
Sbjct 370 CIYRDAKKASEVEDLLQNFKSLISRLEEVDPRKLKHEEKLAFWINVHNALVMHAFLAYGI 429
Query 164 SC----RGALLLRSRCSLLSLWPDVKALDATII----FHTGMLWSRLLKAHERFNAGDET 215
R LLL++ ++ +A+ ++I+ H G W RLL A +F AGDE
Sbjct 430 PQNNVKRVLLLLKAAYNIGGHTISAEAIQSSILGCKMSHPGQ-WLRLLFASRKFKAGDE- 487
Query 216 QRLGSWVGHPQ 226
RL + HP+
Sbjct 488 -RLAYAIDHPE 497
> Hs18482383
Length=610
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 63 PSGFPLLLKQRLHHLSARSKTDEQKRDHMHE 93
P FPLL + R++HLS E+ + MHE
Sbjct 274 PEVFPLLQEARMYHLSGNEIISERTKPRMHE 304
> SPAC4F10.07c
Length=758
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 4/70 (5%)
Query 14 GLKDAKEDAGEHKLLPQVH----SIVKEFLTEDEVAYHKQHVEQHHDKEHKCKPSGFPLL 69
G ++ KE LP V+ SI K ++E+ + H +H + K SGF L
Sbjct 431 GPRERKESFSSRNRLPLVNHPIRSIFKHNVSENPITDHSEHAVYDSEFASKDDLSGFIQL 490
Query 70 LKQRLHHLSA 79
L HHL+A
Sbjct 491 LDSHAHHLNA 500
> At2g23410
Length=290
Score = 30.4 bits (67), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 45/115 (39%), Gaps = 22/115 (19%)
Query 55 HDKEHKCKPSGFPLLLKQRLHHLSARSKTDEQKRDHMHERECPDMALPCCCLYTADLEGH 114
H++E K GFP+ L + + + ++ D +KRD Y +G
Sbjct 7 HEEEFKFIYIGFPVFLLKLIGLIKIKAARDNEKRDE--------------GTYVVREDGL 52
Query 115 LRATWVSNSRF-LSAGLCWVHAVDLTESD-------RLLLGVQLCHEVGLHALAA 161
R + F L W LT S RL+ +LC E+G+H ++A
Sbjct 53 QRELMPRHVAFILDGNRRWAKRAGLTTSQGHEAGAKRLIDIAELCFELGVHTVSA 107
> SPBC336.07
Length=1339
Score = 29.3 bits (64), Expect = 8.6, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 35/79 (44%), Gaps = 3/79 (3%)
Query 26 KLLPQVHSIVKEFLTEDEVAY-HKQHVEQHHDKEHKCKPSGFPLLLKQRLHHLSAR--SK 82
KLLP VHS+ EF D + + H + + + + P G LL ++R + S
Sbjct 440 KLLPHVHSLAGEFSPIDSSLFLNSLHHKGNMESNAEVSPDGMTLLPRKRGRPRKSANISV 499
Query 83 TDEQKRDHMHERECPDMAL 101
T R +E P +A+
Sbjct 500 TSSPIRPSKNENNLPSLAI 518
> Hs13325057
Length=690
Score = 29.3 bits (64), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 25/107 (23%), Positives = 38/107 (35%), Gaps = 17/107 (15%)
Query 157 HALAAAISCRGALLLRSRCSLLS------LWPDVKALDATIIFHTGMLWSRLLKAHERFN 210
H + + G L L + C L W D + T+I + G L L ++
Sbjct 338 HVMGLVVGILGCLDLGATCVLAPKFSTSCFWDDCRQHGVTVILYVGELLRYLCNIPQQPE 397
Query 211 AGDETQRLGSWVGHPQLRASIGTGLPGELWPVVKSVFNIPRILEFYS 257
T RL ++G GL ++W + F RI E Y
Sbjct 398 DRTHTVRL-----------AMGNGLRADVWETFQQRFGPIRIWEVYG 433
Lambda K H
0.322 0.136 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5702305406
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40