bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
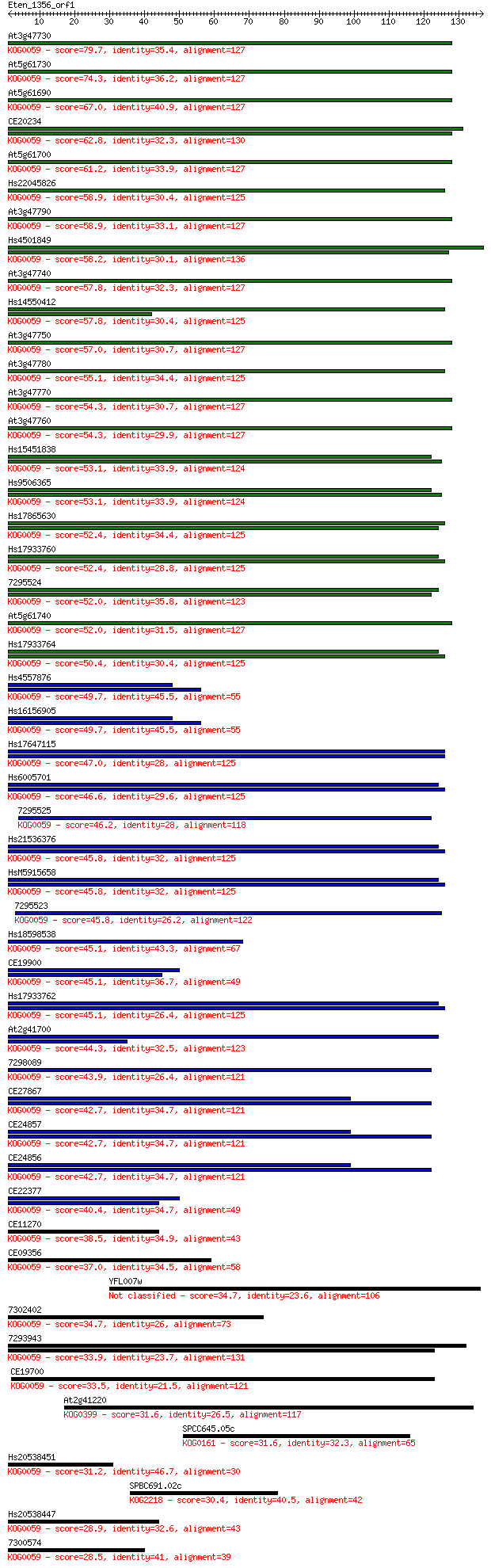
Query= Eten_1356_orf1
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
At3g47730 79.7 1e-15
At5g61730 74.3 6e-14
At5g61690 67.0 9e-12
CE20234 62.8 2e-10
At5g61700 61.2 5e-10
Hs22045826 58.9 2e-09
At3g47790 58.9 2e-09
Hs4501849 58.2 4e-09
At3g47740 57.8 5e-09
Hs14550412 57.8 6e-09
At3g47750 57.0 9e-09
At3g47780 55.1 4e-08
At3g47770 54.3 6e-08
At3g47760 54.3 7e-08
Hs15451838 53.1 1e-07
Hs9506365 53.1 1e-07
Hs17865630 52.4 2e-07
Hs17933760 52.4 3e-07
7295524 52.0 3e-07
At5g61740 52.0 3e-07
Hs17933764 50.4 9e-07
Hs4557876 49.7 2e-06
Hs16156905 49.7 2e-06
Hs17647115 47.0 1e-05
Hs6005701 46.6 1e-05
7295525 46.2 2e-05
Hs21536376 45.8 2e-05
HsM5915658 45.8 2e-05
7295523 45.8 3e-05
Hs18598538 45.1 3e-05
CE19900 45.1 4e-05
Hs17933762 45.1 4e-05
At2g41700 44.3 6e-05
7298089 43.9 8e-05
CE27867 42.7 2e-04
CE24857 42.7 2e-04
CE24856 42.7 2e-04
CE22377 40.4 9e-04
CE11270 38.5 0.003
CE09356 37.0 0.012
YFL007w 34.7 0.049
7302402 34.7 0.050
7293943 33.9 0.083
CE19700 33.5 0.13
At2g41220 31.6 0.42
SPCC645.05c 31.6 0.43
Hs20538451 31.2 0.60
SPBC691.02c 30.4 1.1
Hs20538447 28.9 3.2
7300574 28.5 3.6
> At3g47730
Length=1011
Score = 79.7 bits (195), Expect = 1e-15, Method: Composition-based stats.
Identities = 45/129 (34%), Positives = 71/129 (55%), Gaps = 2/129 (1%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEEAD L DR+ IM G L GT++ LKSRFG+G+I +I F + + + + +
Sbjct 751 MEEADILSDRIGIMAKGRLRCIGTSIRLKSRFGTGFIANISFVESNNHNGEAGSDSREPV 810
Query 61 LQTFEYPPAIKEVSRHE--LRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEE 118
+ F+ +K + ++ + + P L F+EL++R +G+ LG A+LEE
Sbjct 811 KKFFKDHLKVKPIEENKAFMTFVIPHDKENLLTSFFAELQDREEEFGISDIQLGLATLEE 870
Query 119 VFLNVVRIA 127
VFLN+ R A
Sbjct 871 VFLNIARKA 879
> At5g61730
Length=940
Score = 74.3 bits (181), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 46/128 (35%), Positives = 72/128 (56%), Gaps = 6/128 (4%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIF-QKKDEREEQQAKKRLKQ 59
MEEAD L DR+ IM G L GT++ LKSRFG+G++ ++ F + K + + K+ K+
Sbjct 715 MEEADILSDRIGIMAKGRLRCIGTSIRLKSRFGTGFVATVSFIENKKDGAPEPLKRFFKE 774
Query 60 LLQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEV 119
L+ E ++ + + P L F+EL++R + +G+ LG A+LEEV
Sbjct 775 RLKV-----EPTEENKAFMTFVIPHDKEQLLKGFFAELQDRESEFGIADIQLGLATLEEV 829
Query 120 FLNVVRIA 127
FLN+ R A
Sbjct 830 FLNIARRA 837
> At5g61690
Length=954
Score = 67.0 bits (162), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 52/159 (32%), Positives = 73/159 (45%), Gaps = 32/159 (20%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIF--QKKDER-----EEQQA 53
MEEAD L DR+ IM G L GT++ LKSRFG+G++ ++ F K D +
Sbjct 695 MEEADILSDRIGIMAKGRLRCIGTSIRLKSRFGTGFVATVSFIENKNDNNIGVGASHEPL 754
Query 54 KKRLKQLLQ-----------TFEYP--------------PAIKEVSRHELRIISPFCHSF 88
KK K+ L+ TF P P +S +++ C
Sbjct 755 KKFFKEHLKVEPTEENKAFMTFVIPHDKENLLTVQGSQNPIDLHLSIYDIYSSFFLCGLN 814
Query 89 QLPLLFSELENRGALYGVESTVLGFASLEEVFLNVVRIA 127
F EL+NR + +G+ LG A+LEEVFLN+ R A
Sbjct 815 DQQGFFEELQNRESEFGISDIQLGLATLEEVFLNIARQA 853
> CE20234
Length=1802
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 42/132 (31%), Positives = 62/132 (46%), Gaps = 8/132 (6%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEEAD L DR+AIM G L G+ + LK ++G GY ++I++ + + ++
Sbjct 787 MEEADLLGDRIAIMAHGQLECCGSPMFLKQQYGDGYHLTIVYDTTSTPDVSKTTDIIR-- 844
Query 61 LQTFEYPPAIKEVSR--HELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEE 118
EY P S E + H P LF ELE+ G+ S + ++EE
Sbjct 845 ----EYIPEAHVFSYIGQEATYLLSATHRPIFPKLFKELEDHQTQCGITSFGVSITTMEE 900
Query 119 VFLNVVRIATQR 130
VFL V A +R
Sbjct 901 VFLKVGHTADER 912
Score = 50.4 bits (119), Expect = 9e-07, Method: Composition-based stats.
Identities = 40/134 (29%), Positives = 64/134 (47%), Gaps = 21/134 (15%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+E + LC LAIMV G G+ +KSR+GSGY +++ + K+ + ++ K +KQ
Sbjct 1640 MDECEALCTELAIMVYGKFRCYGSCQHIKSRYGSGY--TLLIRLKNRNDAEKTKSTIKQT 1697
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLP-------LLFSELENRGALYGVESTVLGF 113
+ IKE H L++ +F +P LF +LE + L
Sbjct 1698 FRG----SVIKE--EHVLQL------NFDIPRDGDSWSRLFEKLETVSTSLNWDDYSLSQ 1745
Query 114 ASLEEVFLNVVRIA 127
+LE+VF+ R A
Sbjct 1746 TTLEQVFIEFSRDA 1759
> At5g61700
Length=888
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 43/134 (32%), Positives = 66/134 (49%), Gaps = 22/134 (16%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQ-------A 53
MEEA++LCDRL I VDG L G + LKSR+G Y+ ++ K E E ++
Sbjct 763 MEEAEFLCDRLGIFVDGGLQCIGNSKELKSRYGGSYVFTMTTSSKHEEEVERLVESVSPN 822
Query 54 KKRLKQLLQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGF 113
K++ L T ++ E+ + E+RI +F +E A + V + L
Sbjct 823 AKKIYHLAGTQKF-----ELPKQEVRIAE----------VFRAVEKAKANFTVFAWGLAD 867
Query 114 ASLEEVFLNVVRIA 127
+LE+VF+ V R A
Sbjct 868 TTLEDVFIKVARTA 881
> Hs22045826
Length=337
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 67/128 (52%), Gaps = 13/128 (10%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC RLAIMV+G L G+ LK+RFG GY+I++ ++ + +K +
Sbjct 147 MEECEALCTRLAIMVNGRLRCLGSIQHLKNRFGDGYMITV---------RTKSSQSVKDV 197
Query 61 LQTF--EYPPAIKEVSRHELRIISPF-CHSFQLPLLFSELENRGALYGVESTVLGFASLE 117
++ F +P A+ + RH ++ L +FS++E + G+E + +L+
Sbjct 198 VRFFNRNFPEAMLK-ERHHTKVQYQLKSEHISLAQVFSKMEQVSGVLGIEDYSVSQTTLD 256
Query 118 EVFLNVVR 125
VF+N +
Sbjct 257 NVFVNFAK 264
> At3g47790
Length=727
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 66/136 (48%), Gaps = 26/136 (19%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQ-------- 52
MEEA+ LCDR+ I VDG L G LKSR+G Y++++ ++ E+E +Q
Sbjct 609 MEEAEILCDRIGIFVDGSLQCIGNPKELKSRYGGSYVLTVTTSEEHEKEVEQLVHNISTN 668
Query 53 AKKRLKQL-LQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVL 111
AKK + Q FE P + E++I +F LE ++ V + L
Sbjct 669 AKKIYRTAGTQKFELP-------KQEVKIGE----------VFKALEKAKTMFPVVAWGL 711
Query 112 GFASLEEVFLNVVRIA 127
+LE+VF+ V + +
Sbjct 712 ADTTLEDVFIKVAQTS 727
> Hs4501849
Length=1704
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 41/136 (30%), Positives = 70/136 (51%), Gaps = 8/136 (5%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EAD L DR+AIM G L G++L LK ++G+GY ++++ +E + + QL
Sbjct 723 MDEADLLGDRIAIMAKGELQCCGSSLFLKQKYGAGYHMTLV------KEPHCNPEDISQL 776
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVF 120
+ ++ + EL I P + + LF++LE + G+ S ++EEVF
Sbjct 777 VHHHVPNATLESSAGAELSFILPRESTHRFEGLFAKLEKKQKELGIASFGASITTMEEVF 836
Query 121 LNVVRIATQRRSPDLQ 136
L V ++ S D+Q
Sbjct 837 LRVGKLVDS--SMDIQ 850
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/126 (30%), Positives = 57/126 (45%), Gaps = 4/126 (3%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC RLAIMV G G+ LKS+FGSGY + Q + ++E + K L
Sbjct 1574 MEECEALCTRLAIMVQGQFKCLGSPQHLKSKFGSGYSLRAKVQSEGQQEALEEFKAFVDL 1633
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVF 120
+P ++ E + +F LE YGV+ + SLE+VF
Sbjct 1634 ----TFPGSVLEDEHQGMVHYHLPGRDLSWAKVFGILEKAKEKYGVDDYSVSQISLEQVF 1689
Query 121 LNVVRI 126
L+ +
Sbjct 1690 LSFAHL 1695
> At3g47740
Length=925
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 41/136 (30%), Positives = 65/136 (47%), Gaps = 26/136 (19%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDERE----EQQAKKR 56
MEEA++LCDRL I VDG L G LK R+G Y++++ + E++ Q+
Sbjct 800 MEEAEFLCDRLGIFVDGRLQCIGNPKELKGRYGGSYVLTMTTSSEHEKDVEMLVQEVSPN 859
Query 57 LKQLL-----QTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVL 111
+K++ Q FE P + E+RI +F +E + + V + L
Sbjct 860 VKKIYHIAGTQKFEIP-------KDEVRISE----------VFQVVEKAKSNFKVFAWGL 902
Query 112 GFASLEEVFLNVVRIA 127
+LE+VF+ V R A
Sbjct 903 ADTTLEDVFIKVARTA 918
> Hs14550412
Length=2436
Score = 57.8 bits (138), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 67/128 (52%), Gaps = 13/128 (10%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC RLAIMV+G L G+ LK+RFG GY+I++ ++ + +K +
Sbjct 2246 MEECEALCTRLAIMVNGRLRCLGSIQHLKNRFGDGYMITV---------RTKSSQSVKDV 2296
Query 61 LQTF--EYPPAIKEVSRHELRIISPF-CHSFQLPLLFSELENRGALYGVESTVLGFASLE 117
++ F +P A+ + RH ++ L +FS++E + G+E + +L+
Sbjct 2297 VRFFNRNFPEAMLK-ERHHTKVQYQLKSEHISLAQVFSKMEQVSGVLGIEDYSVSQTTLD 2355
Query 118 EVFLNVVR 125
VF+N +
Sbjct 2356 NVFVNFAK 2363
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISII 41
M+EAD L DR+AI+ G L G+ L LK +G GY ++++
Sbjct 1182 MDEADLLGDRIAIISHGKLKCCGSPLFLKGTYGDGYRLTLV 1222
> At3g47750
Length=895
Score = 57.0 bits (136), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 39/134 (29%), Positives = 65/134 (48%), Gaps = 22/134 (16%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQ-------A 53
MEEA++LCDRL I VDG L G LK+R+G Y++++ + E++ +
Sbjct 770 MEEAEFLCDRLGIFVDGRLQCVGNPKELKARYGGSYVLTMTTSSEHEKDVEMLIQDVSPN 829
Query 54 KKRLKQLLQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGF 113
K++ + T ++ E+ + E+RI LF +E + V + L
Sbjct 830 AKKIYHIAGTQKF-----EIPKDEVRIAE----------LFQAVEKAKGNFRVFAWGLAD 874
Query 114 ASLEEVFLNVVRIA 127
+LE+VF+ V R A
Sbjct 875 TTLEDVFIKVARTA 888
> At3g47780
Length=900
Score = 55.1 bits (131), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 43/133 (32%), Positives = 62/133 (46%), Gaps = 24/133 (18%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEEA++LCDRL I VDG L G LK R+G Y +F E +Q ++L
Sbjct 775 MEEAEFLCDRLGIFVDGGLQCIGNPKELKGRYGGSY----VFTMTTSSEHEQNVEKL--- 827
Query 61 LQTFEYPPAIKEVSRHELRIIS-PFCHSFQLPL-------LFSELENRGALYGVESTVLG 112
IK+VS + +I F+LP +F +E + + V + L
Sbjct 828 ---------IKDVSPNAKKIYHIAGTQKFELPKEEVRISEVFQAVEKAKSNFTVFAWGLA 878
Query 113 FASLEEVFLNVVR 125
+LE+VF+ VVR
Sbjct 879 DTTLEDVFIKVVR 891
> At3g47770
Length=722
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 39/136 (28%), Positives = 63/136 (46%), Gaps = 26/136 (19%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDERE----EQQAKKR 56
MEEA++LCDRL I VDG L G LK R+G Y+++I + E++ Q+
Sbjct 603 MEEAEFLCDRLGIFVDGRLQCIGNPKELKGRYGGSYVLTITTSPEHEKDVETLVQEVSSN 662
Query 57 LKQLL-----QTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVL 111
+++ Q FE+P + E+RI +F +EN + V +
Sbjct 663 ARKIYHIAGTQKFEFP-------KEEVRISE----------VFQAVENAKRNFTVFAWGF 705
Query 112 GFASLEEVFLNVVRIA 127
+LE+VF+ V +
Sbjct 706 ADTTLEDVFIKVAKTG 721
> At3g47760
Length=664
Score = 54.3 bits (129), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 38/134 (28%), Positives = 65/134 (48%), Gaps = 22/134 (16%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQ-------A 53
MEEA++LCDRL I VDG L G LK+R+G Y++++ + E++ +
Sbjct 539 MEEAEFLCDRLGIFVDGRLQCVGNPKELKARYGGSYVLTMTTPSEHEKDVEMLVQDVSPN 598
Query 54 KKRLKQLLQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGF 113
K++ + T ++ E+ + E+RI +F +E + V + L
Sbjct 599 AKKIYHIAGTQKF-----EIPKEEVRISE----------VFQAVEKAKDNFRVFAWGLAD 643
Query 114 ASLEEVFLNVVRIA 127
+LE+VF+ V R A
Sbjct 644 TTLEDVFIKVARTA 657
> Hs15451838
Length=2008
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 57/122 (46%), Gaps = 9/122 (7%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC RLAIMV+G G+ LK RF +G+ +++ A+ +
Sbjct 1847 MEECEALCSRLAIMVNGRFRCLGSPQHLKGRFAAGHTLTL--------RVPAARSQPAAA 1898
Query 61 LQTFEYPPA-IKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEV 119
E+P + ++E LR P L +F EL GA +GVE + LEEV
Sbjct 1899 FVAAEFPGSELREAHGGRLRFQLPPGGRCALARVFGELAVHGAEHGVEDFSVSQTMLEEV 1958
Query 120 FL 121
FL
Sbjct 1959 FL 1960
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 46/155 (29%), Positives = 71/155 (45%), Gaps = 36/155 (23%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQK---------------- 44
++EA+ L DR+A++ G L G+ L L+ GSGY ++++ +
Sbjct 860 LDEAELLGDRVAVVAGGRLCCCGSPLFLRRHLGSGYYLTLVKARLPLTTNEKADTDMEGS 919
Query 45 KDEREE-----QQAKKRLKQLLQTFE-YPPAIKEVSR--HELRIISPF--CHSFQLPLLF 94
D R+E Q ++ QLL + + P + V HEL ++ P+ H LF
Sbjct 920 VDTRQEKKNGSQGSRVGTPQLLALVQHWVPGARLVEELPHELVLVLPYTGAHDGSFATLF 979
Query 95 SELENRGA-----LYGVESTVLGFASLEEVFLNVV 124
EL+ R A YG+ T SLEE+FL VV
Sbjct 980 RELDTRLAELRLTGYGISDT-----SLEEIFLKVV 1009
> Hs9506365
Length=2146
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 57/122 (46%), Gaps = 9/122 (7%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC RLAIMV+G G+ LK RF +G+ +++ A+ +
Sbjct 1985 MEECEALCSRLAIMVNGRFRCLGSPQHLKGRFAAGHTLTL--------RVPAARSQPAAA 2036
Query 61 LQTFEYPPA-IKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEV 119
E+P + ++E LR P L +F EL GA +GVE + LEEV
Sbjct 2037 FVAAEFPGSELREAHGGRLRFQLPPGGRCALARVFGELAVHGAEHGVEDFSVSQTMLEEV 2096
Query 120 FL 121
FL
Sbjct 2097 FL 2098
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 46/155 (29%), Positives = 71/155 (45%), Gaps = 36/155 (23%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQK---------------- 44
++EA+ L DR+A++ G L G+ L L+ GSGY ++++ +
Sbjct 998 LDEAELLGDRVAVVAGGRLCCCGSPLFLRRHLGSGYYLTLVKARLPLTTNEKADTDMEGS 1057
Query 45 KDEREE-----QQAKKRLKQLLQTFE-YPPAIKEVSR--HELRIISPF--CHSFQLPLLF 94
D R+E Q ++ QLL + + P + V HEL ++ P+ H LF
Sbjct 1058 VDTRQEKKNGSQGSRVGTPQLLALVQHWVPGARLVEELPHELVLVLPYTGAHDGSFATLF 1117
Query 95 SELENRGA-----LYGVESTVLGFASLEEVFLNVV 124
EL+ R A YG+ T SLEE+FL VV
Sbjct 1118 RELDTRLAELRLTGYGISDT-----SLEEIFLKVV 1147
> Hs17865630
Length=1642
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 66/126 (52%), Gaps = 5/126 (3%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEEA+ +CDR+AIMV G L GT LKS+FG GY + I + KD E + RL++
Sbjct 1493 MEEAEAVCDRVAIMVSGQLRCIGTVQHLKSKFGKGYFLEI--KLKDWIENLEV-DRLQRE 1549
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQ-LPLLFSELENRGALYGVESTVLGFASLEEV 119
+Q + +P A ++ S + Q L F +LE + +E A+LE+V
Sbjct 1550 IQ-YIFPNASRQESFSSILAYKIPKEDVQSLSQSFFKLEEAKHAFAIEEYSFSQATLEQV 1608
Query 120 FLNVVR 125
F+ + +
Sbjct 1609 FVELTK 1614
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/125 (30%), Positives = 68/125 (54%), Gaps = 11/125 (8%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EAD L DR A++ G+L G+++ LKS++G GY +S+ K A + L L
Sbjct 673 MDEADILADRKAVISQGMLKCVGSSMFLKSKWGIGYRLSMYIDK------YCATESLSSL 726
Query 61 LQTFEYPPAIKEVSRHELRIIS--PFCHSFQLPLLFSELENRGALYGVESTVLGFASLEE 118
++ ++ P + +++ +++ PF + LFS L++ L GV S + +LE+
Sbjct 727 VK--QHIPGATLLQQNDQQLVYSLPFKDMDKFSGLFSALDSHSNL-GVISYGVSMTTLED 783
Query 119 VFLNV 123
VFL +
Sbjct 784 VFLKL 788
> Hs17933760
Length=1543
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/125 (28%), Positives = 67/125 (53%), Gaps = 11/125 (8%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EAD L DR + +G L G++L LK ++G GY +S+ R E +++ L
Sbjct 586 MDEADILADRKVFLSNGKLKCAGSSLFLKRKWGIGYHLSL------HRNEMCDTEKITSL 639
Query 61 LQTFEYPPAIKEVSRHELRIIS--PFCHSFQLPLLFSELENRGALYGVESTVLGFASLEE 118
++ ++ P K + E +++ P + + P L+S+L+ + + G+ + + SL E
Sbjct 640 IK--QHIPDAKLTTESEEKLVYSLPLEKTNKFPDLYSDLD-KCSDQGIRNYAVSVTSLNE 696
Query 119 VFLNV 123
VFLN+
Sbjct 697 VFLNL 701
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 38/133 (28%), Positives = 64/133 (48%), Gaps = 22/133 (16%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M EA+ +CDR+A+MV G L G+ LK++FG Y++ I +E Q + ++
Sbjct 1400 MSEAEAVCDRMAMMVSGTLRCIGSIQHLKNKFGRDYLLEI-----KMKEPTQVEALHTEI 1454
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLPL--------LFSELENRGALYGVESTVLG 112
L+ F P A + L +++LP+ F +LE + +E L
Sbjct 1455 LKLF--PQAAWQERYSSLM-------AYKLPVEDVHPLSRAFFKLEAMKQTFNLEEYSLS 1505
Query 113 FASLEEVFLNVVR 125
A+LE+VFL + +
Sbjct 1506 QATLEQVFLELCK 1518
> 7295524
Length=1713
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 44/144 (30%), Positives = 65/144 (45%), Gaps = 27/144 (18%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGY-IIS------------------II 41
M+EAD L DR+AIM DG L +GT+ LK ++GSGY ++S I
Sbjct 705 MDEADVLGDRIAIMCDGELKCQGTSFFLKKQYGSGYRLVSGVQNLFYGRCTYKTCDSLKI 764
Query 42 FQKKDEREEQQAKKRLKQLLQTFEYPPAIKEVSR--HELRIISPFCHSFQLPLLFSELEN 99
K+D+ E + L + Y P +K EL P S + +F +LE
Sbjct 765 CVKRDDCETNEVTALLNK------YIPGLKPECDIGAELSYQLPDSASAKFEEMFGQLEE 818
Query 100 RGALYGVESTVLGFASLEEVFLNV 123
+ + +G S+EEVF+ V
Sbjct 819 QSDELHLNGYGVGITSMEEVFMKV 842
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 41/150 (27%), Positives = 65/150 (43%), Gaps = 37/150 (24%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQ--------- 51
MEE + LC RLAIMV+G G+ LK++F G I+ I ++ E Q
Sbjct 1558 MEECEALCTRLAIMVNGEFKCIGSTQHLKNKFSKGLILKIKVRRNLEALRQARLSGGYAR 1617
Query 52 ---------QAKKR----LKQLLQTFEYPPAIKEVSRHELRIISPFCHSFQLPL------ 92
Q +R +K+ ++T EYP +I + + +F +PL
Sbjct 1618 NPDEQTVPAQMSQRDIDAVKEFVET-EYPNSILQEEYQGIL-------TFYIPLTGVKWS 1669
Query 93 -LFSELENRGALYGVESTVLGFASLEEVFL 121
+F +E+ VE + +LEE+FL
Sbjct 1670 RIFGLMESNRDQLNVEDYSVSQTTLEEIFL 1699
> At5g61740
Length=848
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 40/134 (29%), Positives = 63/134 (47%), Gaps = 22/134 (16%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQ-KKDEREEQQAK----- 54
MEEA++LCDRL I VDG L G LK R+G Y+ ++ + +E+ E+ K
Sbjct 723 MEEAEFLCDRLGIFVDGGLQCVGNPKELKGRYGGSYVFTMTTSVEHEEKVERMVKHISPN 782
Query 55 -KRLKQLLQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGF 113
KR+ L T ++ E+ + E+ I +F +E + + V + L
Sbjct 783 SKRVYHLAGTQKF-----EIPKQEVMIAD----------VFFMVEKVKSKFTVFAWGLAD 827
Query 114 ASLEEVFLNVVRIA 127
+LE+VF V A
Sbjct 828 TTLEDVFFKVATTA 841
> Hs17933764
Length=1617
Score = 50.4 bits (119), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 64/124 (51%), Gaps = 9/124 (7%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EAD L DR IM +G L G+++ LK R+G GY +S+ R E +++
Sbjct 673 MDEADILADRKVIMSNGRLKCAGSSMFLKRRWGLGYHLSL------HRNEICNPEQITSF 726
Query 61 LQTFEYPPA-IKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEV 119
+ T P A +K ++ +L P + P LFS+L+ + + GV + ++L EV
Sbjct 727 I-THHIPDAKLKTENKEKLVYTLPLERTNTFPDLFSDLD-KCSDQGVTGYDISMSTLNEV 784
Query 120 FLNV 123
F+ +
Sbjct 785 FMKL 788
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 58/125 (46%), Gaps = 6/125 (4%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
+ EA+ LCDR+AIMV G L G+ LK++ G YI+ + +E Q ++
Sbjct 1473 LAEAEALCDRVAIMVSGRLRCIGSIQHLKNKLGKDYILEL-----KVKETSQVTLVHTEI 1527
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVF 120
L+ F A +E L P + L F +LE + +E L +LE+VF
Sbjct 1528 LKLFP-QAAGQERYSSLLTYKLPVADVYPLSQTFHKLEAVKHNFNLEEYSLSQCTLEKVF 1586
Query 121 LNVVR 125
L + +
Sbjct 1587 LELSK 1591
> Hs4557876
Length=2273
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 31/48 (64%), Gaps = 1/48 (2%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISI-IFQKKDE 47
MEE + LC RLAIMV G GT LKS+FG GYI+++ I KD+
Sbjct 2130 MEECEALCTRLAIMVKGAFRCMGTIQHLKSKFGDGYIVTMKIKSPKDD 2177
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKK 55
M+EAD+ DR+AI+ G L GT L LK+ FG+G ++++ + K+ + +++ +
Sbjct 1120 MDEADHQGDRIAIIAQGRLYCSGTPLFLKNCFGTGLYLTLVRKMKNIQSQRKGSE 1174
> Hs16156905
Length=2273
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 31/48 (64%), Gaps = 1/48 (2%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISI-IFQKKDE 47
MEE + LC RLAIMV G GT LKS+FG GYI+++ I KD+
Sbjct 2130 MEECEALCTRLAIMVKGAFRCMGTIQHLKSKFGDGYIVTMKIKSPKDD 2177
Score = 43.9 bits (102), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKK 55
M+EAD L DR+AI+ G L GT L LK+ FG+G ++++ + K+ + +++ +
Sbjct 1120 MDEADLLGDRIAIIAQGRLYCSGTPLFLKNCFGTGLYLTLVRKMKNIQSQRKGSE 1174
> Hs17647115
Length=2277
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 58/126 (46%), Gaps = 8/126 (6%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC RLAIMV+G G+ +KSRFG G+ + + K+ + + + QL
Sbjct 2131 MEECEALCTRLAIMVNGKFQCIGSLQHIKSRFGRGFTVKV--HLKNNKVTMETLTKFMQL 2188
Query 61 LQTFEYPPA-IKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEV 119
+P +K+ L P + +F LE + + ++ +LEEV
Sbjct 2189 ----HFPKTYLKDQHLSMLEYHVPVTAG-GVANIFDLLETNKTALNITNFLVSQTTLEEV 2243
Query 120 FLNVVR 125
F+N +
Sbjct 2244 FINFAK 2249
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 60/135 (44%), Gaps = 15/135 (11%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRL--K 58
++EA+ L DR+A + G L G+ LK FG GY +++ +K +
Sbjct 1219 LDEAEVLSDRIAFLEQGGLRCCGSPFYLKEAFGDGYHLTLTKKKSPNLNANAVCDTMAVT 1278
Query 59 QLLQTFEYPPAIKEVSRHEL-RIISPFCHSFQ---LPLLFSELENRGAL----YGVESTV 110
++Q+ +KE EL ++ PF L LL + G L YG+ T
Sbjct 1279 AMIQSHLPEAYLKEDIGGELVYVLPPFSTKVSGAYLSLLRALDNGMGDLNIGCYGISDT- 1337
Query 111 LGFASLEEVFLNVVR 125
++EEVFLN+ +
Sbjct 1338 ----TVEEVFLNLTK 1348
> Hs6005701
Length=1581
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 64/125 (51%), Gaps = 11/125 (8%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EAD L DR + G L G++L LK ++G GY +S+ Q + E+ +KQ
Sbjct 635 MDEADILADRKVFLSQGKLKCAGSSLFLKKKWGIGYHLSL--QLNEICVEENITSLVKQ- 691
Query 61 LQTFEYPPAIKEVSRHELRIIS--PFCHSFQLPLLFSELENRGALYGVESTVLGFASLEE 118
+ P K ++ E ++I P + + P L+ +L++ L G+E+ + +L E
Sbjct 692 -----HIPDAKLSAKSEGKLIYTLPLERTNKFPELYKDLDSYPDL-GIENYGVSMTTLNE 745
Query 119 VFLNV 123
VFL +
Sbjct 746 VFLKL 750
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 62/126 (49%), Gaps = 8/126 (6%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M EA+ +CDR+AIMV G L G+ LKS+FG Y++ + + + E A ++
Sbjct 1438 MAEAEAVCDRVAIMVSGRLRCIGSIQHLKSKFGKDYLLEMKVKNLAQVEPLHA-----EI 1492
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQ-LPLLFSELENRGALYGVESTVLGFASLEEV 119
L+ F P A ++ L + Q L F +LE + +E L ++LE+V
Sbjct 1493 LRLF--PQAARQERYSSLMVYKLPVEDVQPLAQAFFKLEKVKQSFDLEEYSLSQSTLEQV 1550
Query 120 FLNVVR 125
FL + +
Sbjct 1551 FLELSK 1556
> 7295525
Length=1197
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 33/118 (27%), Positives = 59/118 (50%), Gaps = 6/118 (5%)
Query 4 ADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQLLQT 63
A+ + DR+AIM +G L GT LK+ +G GY ++ + K +R+E L ++ +
Sbjct 363 AENIADRMAIMSNGELKCTGTKPFLKNMYGHGYRLTCVKGKNYKRDE------LFGMMNS 416
Query 64 FEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVFL 121
+ +I+ +++ + Q P+L +LE GV S + S+EE+FL
Sbjct 417 YMPNMSIERDIGYKVTFVLENKFEDQFPMLIDDLEENMQQLGVVSFRIRDTSMEEIFL 474
> Hs21536376
Length=2261
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 73/161 (45%), Gaps = 38/161 (23%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISII------------------- 41
M+EAD L DR+AI+ G L G++L LK++ G+GY ++++
Sbjct 1091 MDEADVLGDRIAIISHGKLCCVGSSLFLKNQLGTGYYLTLVKKDVESSLSSCRNSSSTVS 1150
Query 42 -FQKKDEREEQQAKKRLKQLLQTFEYPPAIKEVS----RH--ELRIISPFCH--SFQLPL 92
+K+D + + L ++ + +S +H E R++ H ++ LP
Sbjct 1151 YLKKEDSVSQSSSDAGLGSDHESDTLTIDVSAISNLIRKHVSEARLVEDIGHELTYVLPY 1210
Query 93 ----------LFSELENRGALYGVESTVLGFASLEEVFLNV 123
LF E+++R + G+ S + +LEE+FL V
Sbjct 1211 EAAKEGAFVELFHEIDDRLSDLGISSYGISETTLEEIFLKV 1251
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 60/128 (46%), Gaps = 12/128 (9%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC R+AIMV+G G+ LK+RFG GY I + + LK +
Sbjct 2104 MEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGDGYTIVVRIAG--------SNPDLKPV 2155
Query 61 LQTF--EYPPAI-KEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLE 117
F +P ++ KE R+ L+ P S L +FS L +E + +L+
Sbjct 2156 QDFFGLAFPGSVLKEKHRNMLQYQLPSSLS-SLARIFSILSQSKKRLHIEDYSVSQTTLD 2214
Query 118 EVFLNVVR 125
+VF+N +
Sbjct 2215 QVFVNFAK 2222
> HsM5915658
Length=2201
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 73/161 (45%), Gaps = 38/161 (23%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISII------------------- 41
M+EAD L DR+AI+ G L G++L LK++ G+GY ++++
Sbjct 1031 MDEADVLGDRIAIISHGKLCCVGSSLFLKNQLGTGYYLTLVKKDVESSLSSCRNSSSTVS 1090
Query 42 -FQKKDEREEQQAKKRLKQLLQTFEYPPAIKEVS----RH--ELRIISPFCH--SFQLPL 92
+K+D + + L ++ + +S +H E R++ H ++ LP
Sbjct 1091 YLKKEDSVSQSSSDAGLGSDHESDTLTIDVSAISNLIRKHVSEARLVEDIGHELTYVLPY 1150
Query 93 ----------LFSELENRGALYGVESTVLGFASLEEVFLNV 123
LF E+++R + G+ S + +LEE+FL V
Sbjct 1151 EAAKEGAFVELFHEIDDRLSDLGISSYGISETTLEEIFLKV 1191
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 60/128 (46%), Gaps = 12/128 (9%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEE + LC R+AIMV+G G+ LK+RFG GY I + + LK +
Sbjct 2044 MEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGDGYTIVVRIAG--------SNPDLKPV 2095
Query 61 LQTF--EYPPAI-KEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLE 117
F +P ++ KE R+ L+ P S L +FS L +E + +L+
Sbjct 2096 QDFFGLAFPGSVPKEKHRNMLQYQLPSSLS-SLARIFSILSQSKKRLHIEDYSVSQTTLD 2154
Query 118 EVFLNVVR 125
+VF+N +
Sbjct 2155 QVFVNFAK 2162
> 7295523
Length=1511
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 59/122 (48%), Gaps = 6/122 (4%)
Query 3 EADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQLLQ 62
EA+++ DR+ I+ GVL + GT L+S+F S + ++ KK +Q + Q +Q
Sbjct 677 EAEHVADRIGILSMGVLEASGTPFFLRSKFSSS--VDLVIIKKPHVPDQPITDYINQFMQ 734
Query 63 TFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVFLN 122
E I + + L P + +L L LE + G+E+ + A L ++++
Sbjct 735 NIEPENEIGDSLTYRL----PVIYRPRLQKLLIHLEIDRKMLGIENVRVVGAELSDIYMT 790
Query 123 VV 124
+V
Sbjct 791 LV 792
> Hs18598538
Length=222
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 41/75 (54%), Gaps = 8/75 (10%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISI-IFQKKDEREEQ-------Q 52
MEE D LC LAIMV G G+ LKS+FG+ YI+ + +F +E +EQ
Sbjct 136 MEECDALCTSLAIMVQGKFTCLGSPQHLKSKFGNIYILKVKVFGILEEAKEQFDLEDYSV 195
Query 53 AKKRLKQLLQTFEYP 67
++ L+Q+ TF P
Sbjct 196 SQITLEQVFLTFANP 210
> CE19900
Length=1431
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 36/49 (73%), Gaps = 0/49 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDERE 49
MEE + LC R+ I+ +G + + GT+ +LKS++G+ Y++++I K ++R+
Sbjct 1313 MEECEALCTRIGILRNGEMIALGTSQSLKSQYGNTYMMTLILNKLEDRK 1361
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQK 44
M+EA+ L D + +M G +A+ G+ LK ++GSG +++I+F+
Sbjct 637 MDEAEKLGDWIFVMSHGKMAASGSNHYLKQKYGSGMLLTIVFKN 680
> Hs17933762
Length=1624
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/123 (26%), Positives = 61/123 (49%), Gaps = 7/123 (5%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
++EAD L DR + +G L G++L LK ++G GY +S+ ++ + E + L
Sbjct 676 IDEADILADRKVFISNGKLKCAGSSLFLKKKWGIGYHLSLHLNERCDPES------ITSL 729
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVF 120
++ + S +L I P + + P L+ +L+ R + G+E + +L EVF
Sbjct 730 VKQHISDAKLTAQSEEKLVYILPLERTNKFPELYRDLD-RCSNQGIEDYGVSITTLNEVF 788
Query 121 LNV 123
L +
Sbjct 789 LKL 791
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 62/126 (49%), Gaps = 8/126 (6%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M EA+ +CDR+AIMV G L G+ LKS+FG Y++ + + + E A ++
Sbjct 1481 MAEAEAVCDRVAIMVSGRLRCIGSIQHLKSKFGKDYLLEMKLKNLAQMEPLHA-----EI 1535
Query 61 LQTFEYPPAIKEVSRHELRIIS-PFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEV 119
L+ F P A ++ L + P L F +LE + +E L ++LE+V
Sbjct 1536 LRLF--PQAAQQERFSSLMVYKLPVEDVRPLSQAFFKLEIVKQSFDLEEYSLSQSTLEQV 1593
Query 120 FLNVVR 125
FL + +
Sbjct 1594 FLELSK 1599
> At2g41700
Length=1850
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 40/136 (29%), Positives = 62/136 (45%), Gaps = 28/136 (20%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EA+ L DR+ IM +G L G+++ LK +G GY ++++ R
Sbjct 676 MDEAEELGDRIGIMANGSLKCCGSSIFLKHHYGVGYTLTLVKTSPTVSVAAHIVHR---- 731
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLPL--------LFSELE----NRGALY-GVE 107
+ P+ VS I SF+LPL +F E+E N + Y G++
Sbjct 732 -----HIPSATCVSEVGNEI------SFKLPLASLPCFENMFREIESCMKNSDSDYPGIQ 780
Query 108 STVLGFASLEEVFLNV 123
S + +LEEVFL V
Sbjct 781 SYGISVTTLEEVFLRV 796
Score = 34.7 bits (78), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGS 34
M EA LC R+ IMV G L G+ LK+R+G+
Sbjct 1613 MNEAQALCTRIGIMVGGRLRCIGSPQHLKTRYGN 1646
> 7298089
Length=324
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 54/121 (44%), Gaps = 5/121 (4%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
MEEA+ L D + I+ +G L S G+ L LK + G GY + + RE + + ++
Sbjct 208 MEEAEVLGDTICILANGKLQSIGSPLELKRKSGIGYRLKLEINDFTSREVE-----IMEI 262
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVF 120
+ F + V + I P+ + + LE G+ + + SLE+VF
Sbjct 263 IHHFVPTARVLNVVNPTVYICLPYAYKNCFAQMLYRLETESKELGIHTISMTDTSLEDVF 322
Query 121 L 121
L
Sbjct 323 L 323
> CE27867
Length=1704
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 12/103 (11%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EA+ L D + IM G L + GT LK +FG+GY+++++ + KR +
Sbjct 737 MDEAERLGDWVFIMSHGKLVASGTNQYLKQKFGTGYLLTVVL-------DHNGDKRKMAV 789
Query 61 LQTFEYPPAIKEVSRHELR-----IISPFCHSFQLPLLFSELE 98
+ T +KE R E+ II P + LF LE
Sbjct 790 ILTDVCTHYVKEAERGEMHGQQIEIILPEARKKEFVPLFQALE 832
Score = 40.8 bits (94), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 35/129 (27%), Positives = 65/129 (50%), Gaps = 19/129 (14%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+E + LC R+A++ G L + G++ LKS +G+ Y +++ + ++R+ + QL
Sbjct 1563 MDECEALCSRIAVLNRGSLIAIGSSQELKSLYGNNYTMTLSLYEPNQRD------MVVQL 1616
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLP-----LLFSELENRGAL---YGVESTVLG 112
+QT +K S ++ + +Q+P ++ E AL GV+ +L
Sbjct 1617 VQTRLPNSVLKTTSTNKTLNL-----KWQIPKEKEDCWSAKFEMVQALAKDLGVKDFILA 1671
Query 113 FASLEEVFL 121
+SLEE FL
Sbjct 1672 QSSLEETFL 1680
> CE24857
Length=1691
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 12/103 (11%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EA+ L D + IM G L + GT LK +FG+GY+++++ + KR +
Sbjct 724 MDEAERLGDWVFIMSHGKLVASGTNQYLKQKFGTGYLLTVVL-------DHNGDKRKMAV 776
Query 61 LQTFEYPPAIKEVSRHELR-----IISPFCHSFQLPLLFSELE 98
+ T +KE R E+ II P + LF LE
Sbjct 777 ILTDVCTHYVKEAERGEMHGQQIEIILPEARKKEFVPLFQALE 819
Score = 40.4 bits (93), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 35/129 (27%), Positives = 65/129 (50%), Gaps = 19/129 (14%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+E + LC R+A++ G L + G++ LKS +G+ Y +++ + ++R+ + QL
Sbjct 1550 MDECEALCSRIAVLNRGSLIAIGSSQELKSLYGNNYTMTLSLYEPNQRD------MVVQL 1603
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLP-----LLFSELENRGAL---YGVESTVLG 112
+QT +K S ++ + +Q+P ++ E AL GV+ +L
Sbjct 1604 VQTRLPNSVLKTTSTNKTLNL-----KWQIPKEKEDCWSAKFEMVQALAKDLGVKDFILA 1658
Query 113 FASLEEVFL 121
+SLEE FL
Sbjct 1659 QSSLEETFL 1667
> CE24856
Length=1689
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 12/103 (11%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+EA+ L D + IM G L + GT LK +FG+GY+++++ + KR +
Sbjct 724 MDEAERLGDWVFIMSHGKLVASGTNQYLKQKFGTGYLLTVVL-------DHNGDKRKMAV 776
Query 61 LQTFEYPPAIKEVSRHELR-----IISPFCHSFQLPLLFSELE 98
+ T +KE R E+ II P + LF LE
Sbjct 777 ILTDVCTHYVKEAERGEMHGQQIEIILPEARKKEFVPLFQALE 819
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/129 (27%), Positives = 65/129 (50%), Gaps = 19/129 (14%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+E + LC R+A++ G L + G++ LKS +G+ Y +++ + ++R+ + QL
Sbjct 1548 MDECEALCSRIAVLNRGSLIAIGSSQELKSLYGNNYTMTLSLYEPNQRD------MVVQL 1601
Query 61 LQTFEYPPAIKEVSRHELRIISPFCHSFQLP-----LLFSELENRGAL---YGVESTVLG 112
+QT +K S ++ + +Q+P ++ E AL GV+ +L
Sbjct 1602 VQTRLPNSVLKTTSTNKTLNL-----KWQIPKEKEDCWSAKFEMVQALAKDLGVKDFILA 1656
Query 113 FASLEEVFL 121
+SLEE FL
Sbjct 1657 QSSLEETFL 1665
> CE22377
Length=1564
Score = 40.4 bits (93), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDERE 49
MEE + LC R+ I+ G + + GT+ +LKS++G+ Y++++I ++ E
Sbjct 1434 MEECEALCTRIGILRKGEMIALGTSQSLKSQYGNTYMMTLILNSLEDLE 1482
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQ 43
M+EA+ L D + +M G +A+ G+ LK ++G G +++++F+
Sbjct 642 MDEAEKLGDWIFVMSHGKMAASGSKHYLKQKYGGGMLLTLVFK 684
> CE11270
Length=260
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 30/43 (69%), Gaps = 0/43 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQ 43
MEE + LC R+ I+ G + + GT+ +LKS++G+ Y+++++
Sbjct 134 MEECEALCTRIGILHRGEMIALGTSQSLKSQYGNTYMMTLVLN 176
> CE09356
Length=1447
Score = 37.0 bits (84), Expect = 0.012, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLK 58
MEE + LC R+AIM G + G LKS++G G ++++ K + +E R K
Sbjct 1354 MEECEALCTRIAIMDRGRIRCLGGKQHLKSKYGKGSMLTMKMGKDENAKEIAGIMRSK 1411
> YFL007w
Length=1804
Score = 34.7 bits (78), Expect = 0.049, Method: Composition-based stats.
Identities = 25/107 (23%), Positives = 51/107 (47%), Gaps = 3/107 (2%)
Query 30 SRFGSGYIISIIFQKKDEREEQQAKKRLKQLLQTFEYPPAIKEVSRHELRIISPFCHSFQ 89
SR ++ +I KKD E+ L + ++TF +P ++ + + F S+
Sbjct 489 SRTVKPFVYAINGSKKDRFFEKLVS--LAKAIETFIHPSNNGFWTKPNAKFVHAFIKSYH 546
Query 90 LPLLFSE-LENRGALYGVESTVLGFASLEEVFLNVVRIATQRRSPDL 135
+ + E + RG G+ T + E+FLN++ + +Q ++PD+
Sbjct 547 GRVKYEEDICARGVTNGICLTSFCHEEIVEIFLNIISLGSQNKNPDI 593
> 7302402
Length=1382
Score = 34.7 bits (78), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 5/73 (6%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
+EE ++LC + IM G L G+ LK RF G + + + E ++++
Sbjct 1187 LEECEFLCTNVGIMDHGSLLCYGSLSRLKHRFNMGIFVKVKMGTRAEMDDER-----DNW 1241
Query 61 LQTFEYPPAIKEV 73
+Q PP E+
Sbjct 1242 MQITMMPPNDSEM 1254
> 7293943
Length=1463
Score = 33.9 bits (76), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 56/132 (42%), Gaps = 6/132 (4%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
M+E + LC + I+VDG + + G+ +K++ G + ++ + + K +
Sbjct 1240 MDEINALCSKSVILVDGSIYAMGSVQHVKNKIAKGMTLKLVVNVQPDNMVAMLTKIEDDI 1299
Query 61 LQTFEYPPA-IKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEV 119
T YP A +KE R+ +F +E + + +E L SLE+
Sbjct 1300 YMT--YPNAELKEKYEFSGRLTFQISKDTSWSEIFEYVEGNRSSWHLEDYSLSQPSLEDA 1357
Query 120 FLNVVRIATQRR 131
F IA +RR
Sbjct 1358 F---EEIAEERR 1366
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 36/135 (26%), Positives = 62/135 (45%), Gaps = 32/135 (23%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQL 60
+++ + L DR+ I+ DG L G+ LK + + +I+ +K+ + E +L L
Sbjct 528 LDDGEVLGDRVVIISDGQLRCIGSLPFLKKQVDASCLITCEARKRCDLE------KLTSL 581
Query 61 LQTFEYPPAIKEVSRHELRIISPFC-----HSFQLPL--------LFSELENRGALYGVE 107
+SRH + I PF ++LPL LF +LE++ + GV
Sbjct 582 ------------ISRH-VGTIQPFSIKGRDVCYKLPLSKSKYFSSLFRDLESQMNILGVR 628
Query 108 STVLGFASLEEVFLN 122
L SLEE+F++
Sbjct 629 GFSLSSVSLEEIFMS 643
> CE19700
Length=1429
Score = 33.5 bits (75), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 26/124 (20%), Positives = 54/124 (43%), Gaps = 5/124 (4%)
Query 2 EEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQLL 61
EE + + D++ +M +G + G+ A K S + I I + ++Q K +K L
Sbjct 554 EETEAISDKVVLMSEGYVVLNGSCEAFKHSINSVFEIRI--WPNNRFTDEQIKGMMKTLT 611
Query 62 ---QTFEYPPAIKEVSRHELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEE 118
+ E ++R+ P + +PL+ EL+ + +E LG ++ +
Sbjct 612 LGDNQMKNDARFFETPNGKIRVTLPILYRRSIPLILRELDAVHEKFRIEFYELGKPNIHD 671
Query 119 VFLN 122
++ N
Sbjct 672 IYAN 675
> At2g41220
Length=1629
Score = 31.6 bits (70), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 49/117 (41%), Gaps = 15/117 (12%)
Query 17 GVLASEGTALALKSRFGSGYIISIIFQKKDEREEQQAKKRLKQLLQTFEYPPAIKEVSRH 76
GVL + + + +K R G G +IS+ + E + KKR+ Y P K VS +
Sbjct 481 GVLPMDESKVTMKGRLGPGMMISVDLENGQVYENTEVKKRVAS------YNPYGKWVSEN 534
Query 77 ELRIISPFCHSFQLPLLFSELENRGALYGVESTVLGFASLEEVFLNVVRIATQRRSP 133
LR + P + L E R +G S E+V + + +A Q + P
Sbjct 535 -LRNLKPSNYLSSAILETDETLRRQQAFGYSS--------EDVQMVIESMAAQGKEP 582
> SPCC645.05c
Length=1526
Score = 31.6 bits (70), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 10/69 (14%)
Query 51 QQAKKRLKQLLQTFEYPPAIKEVSRHELRIISPF----CHSFQLPLLFSELENRGALYGV 106
Q+ K++L QL+ F H +R I P H+F PL+ +L G L G+
Sbjct 628 QRHKEQLNQLMNQF------NSTQPHFIRCIVPNEEKKMHTFNRPLVLGQLRCNGVLEGI 681
Query 107 ESTVLGFAS 115
T GF +
Sbjct 682 RITRAGFPN 690
> Hs20538451
Length=1224
Score = 31.2 bits (69), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKS 30
MEE + LC RLAIMV+G G+ +K+
Sbjct 874 MEECEALCTRLAIMVNGSFKCLGSPQHIKN 903
> SPBC691.02c
Length=678
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 28/46 (60%), Gaps = 4/46 (8%)
Query 36 YIISIIFQKKDEREEQQAKKRLKQLLQT-FEYP---PAIKEVSRHE 77
YI ++FQKK+ +++ K L + L+T F +P P +KE S+ E
Sbjct 134 YIAELLFQKKEREVKEKWKNELTEKLKTLFNWPAINPNLKESSKFE 179
> Hs20538447
Length=4273
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIISIIFQ 43
++EA+ L DR+A++ G L G LK +G G +++ Q
Sbjct 4097 LDEAEALSDRVAVLQHGRLRCCGPPFCLKEAYGQGLRLTLTRQ 4139
> 7300574
Length=494
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 1 MEEADYLCDRLAIMVDGVLASEGTALALKSRFGSGYIIS 39
+EE + L DR+AIM DG + GT LK + Y +S
Sbjct 221 LEEGEILADRIAIMNDGQILCYGTLGYLKQLPFTSYTLS 259
Lambda K H
0.322 0.137 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1425342594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40