bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1361_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
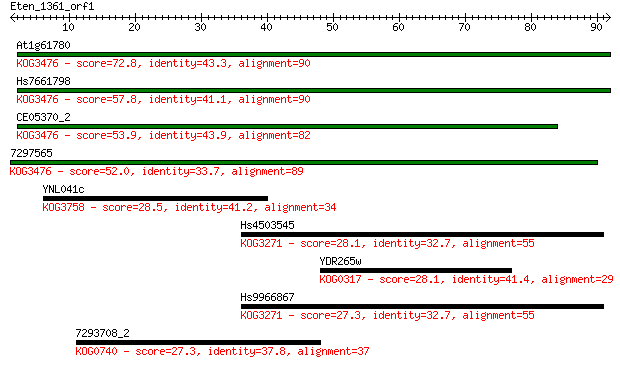
At1g61780 72.8 1e-13
Hs7661798 57.8 5e-09
CE05370_2 53.9 8e-08
7297565 52.0 3e-07
YNL041c 28.5 3.2
Hs4503545 28.1 4.3
YDR265w 28.1 4.6
Hs9966867 27.3 6.9
7293708_2 27.3 7.2
> At1g61780
Length=98
Score = 72.8 bits (177), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 52/98 (53%), Gaps = 8/98 (8%)
Query 2 MPCDKCEAKFTKLVTPD--------VKEGSKRIVGVNKLIEKATKKDKLVIVGSKCKICK 53
M CDKCE K +K++ PD V EG R + NKL+ K + +KC ICK
Sbjct 1 MVCDKCEKKLSKVIVPDKWKDGARNVTEGGGRKINENKLLSKKNRWSPYSTCTTKCMICK 60
Query 54 TALHMKGKYCAPCAHVNGRCWMCGKRIVDVSKHNMSLV 91
+H GKYC CA+ G C MCGK+++D + S V
Sbjct 61 QQVHQDGKYCHTCAYSKGVCAMCGKQVLDTKMYKQSNV 98
> Hs7661798
Length=101
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 37/101 (36%), Positives = 55/101 (54%), Gaps = 11/101 (10%)
Query 2 MPCDKCEAKFTKLVTPDV-KEGSKRIV--GVNKLIEK---ATKKDKLVIVG----SKCKI 51
M C+KCE K ++TPD K+G++ G KL E +KK + G S C+I
Sbjct 1 MVCEKCEKKLGTVITPDTWKDGARNTTESGGRKLNENKALTSKKARFDPYGKNKFSTCRI 60
Query 52 CKTALHMKGK-YCAPCAHVNGRCWMCGKRIVDVSKHNMSLV 91
CK+++H G YC CA+ G C MCGK+++D + + V
Sbjct 61 CKSSVHQPGSHYCQGCAYKKGICAMCGKKVLDTKNYKQTSV 101
> CE05370_2
Length=99
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 47/94 (50%), Gaps = 15/94 (15%)
Query 2 MPCDKCEAKFTKLVTPDV-----------KEGSKRIVGVNKLIEKATKKDKLVIVGSKCK 50
M C CE K TK+V D G K + N+LI + K IVG+KCK
Sbjct 1 MVCGDCEKKLTKIVGVDPYRNKKVNRNADGSGPKTVTTKNRLIGV---QKKATIVGAKCK 57
Query 51 ICKTALHMKGK-YCAPCAHVNGRCWMCGKRIVDV 83
+CK +H G YC+ CA+ G C MCGK+I +
Sbjct 58 LCKMLIHQPGSHYCSTCAYQKGICAMCGKKIQNT 91
> 7297565
Length=116
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 51/96 (53%), Gaps = 7/96 (7%)
Query 1 EMPCDKCEAKFTKLVTPDVKEGSKRIVGVNKLIEK---ATKKDKLVIVGS---KCKICKT 54
+M C+KCEAK +K+ P+ S G K+ E ++ +++ +G+ C+IC+
Sbjct 19 KMVCEKCEAKLSKVSAPNPWRTSTAPAGGRKINENKALSSARERYNPIGTALPPCRICRQ 78
Query 55 ALHMKGK-YCAPCAHVNGRCWMCGKRIVDVSKHNMS 89
+H G YC CA+ C MCGK+I++ + S
Sbjct 79 KVHQMGSHYCQACAYKKAICAMCGKKIMNTKNYKQS 114
> YNL041c
Length=839
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 22/34 (64%), Gaps = 1/34 (2%)
Query 6 KCEAKFTKLVTPDVKEGSKRIVGVNKLIEKATKK 39
K E+ F L P++ G+ I+GVN+++EK KK
Sbjct 314 KDESSFL-LTLPNLNAGNALIMGVNEILEKTNKK 346
> Hs4503545
Length=154
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 3/58 (5%)
Query 36 ATKKDKLVIV-GSKCKICKTALHMKGKYCAPCAHVNGRCWMCGKRIVDV--SKHNMSL 90
A +K+ V++ G CKI + + GK+ H+ G GK+ D+ S HNM +
Sbjct 24 ALRKNGFVVLKGRPCKIVEMSTSKTGKHGHAKVHLVGIDIFTGKKYEDICPSTHNMDV 81
> YDR265w
Length=337
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 16/29 (55%), Gaps = 4/29 (13%)
Query 48 KCKICKTALHMKGKYCAPCAHVNGRCWMC 76
KC +C ++M CAPC H+ CW C
Sbjct 285 KCILC--LMNMSDPSCAPCGHL--FCWSC 309
> Hs9966867
Length=153
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 3/58 (5%)
Query 36 ATKKDKLVIV-GSKCKICKTALHMKGKYCAPCAHVNGRCWMCGKRIVDV--SKHNMSL 90
A +K+ V++ G CKI + + GK+ H+ G GK+ D+ S HNM +
Sbjct 24 ALRKNGFVVLKGRPCKIVEMSTSKTGKHGHAKVHLVGIDIFTGKKYEDICPSTHNMDV 81
> 7293708_2
Length=332
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 3/40 (7%)
Query 11 FTKLVTPDVKEGSKRIVGVN---KLIEKATKKDKLVIVGS 47
F K V P+ E + R++G + K+++ K DK+V+VG+
Sbjct 150 FWKKVPPEAVEINPRLLGTSLASKILKPLKKNDKIVLVGT 189
Lambda K H
0.322 0.135 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40