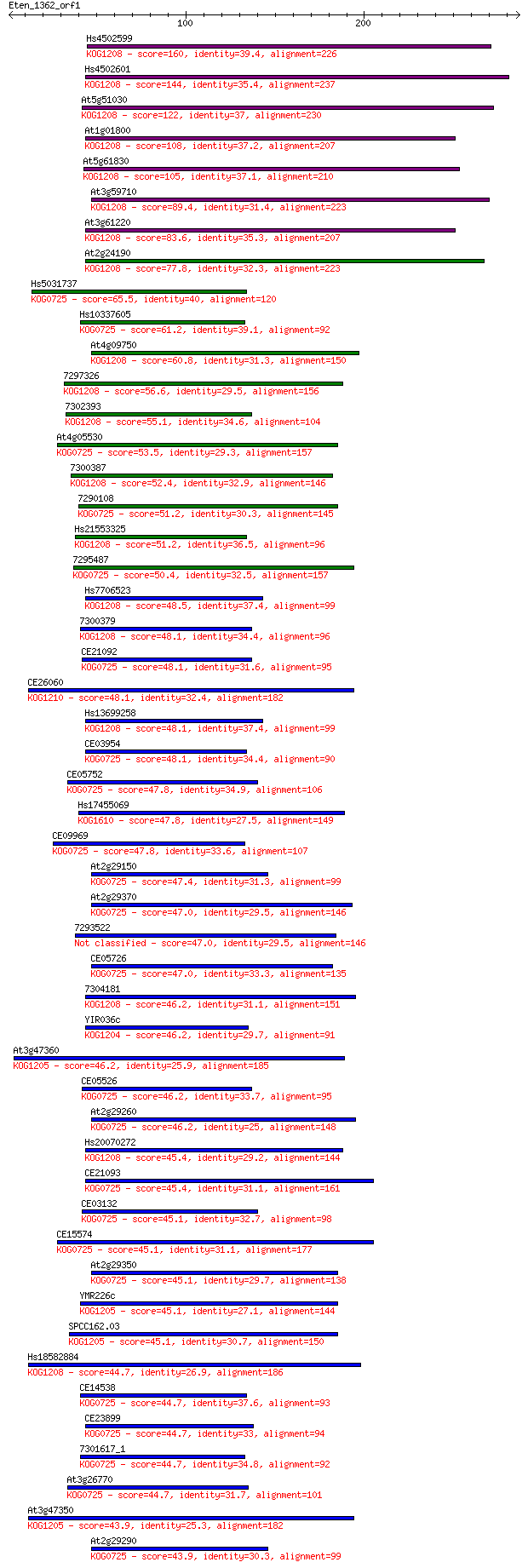
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1362_orf1
Length=286
Score E
Sequences producing significant alignments: (Bits) Value
Hs4502599 160 4e-39
Hs4502601 144 2e-34
At5g51030 122 9e-28
At1g01800 108 1e-23
At5g61830 105 1e-22
At3g59710 89.4 7e-18
At3g61220 83.6 5e-16
At2g24190 77.8 3e-14
Hs5031737 65.5 1e-10
Hs10337605 61.2 2e-09
At4g09750 60.8 3e-09
7297326 56.6 5e-08
7302393 55.1 2e-07
At4g05530 53.5 5e-07
7300387 52.4 1e-06
7290108 51.2 2e-06
Hs21553325 51.2 2e-06
7295487 50.4 4e-06
Hs7706523 48.5 2e-05
7300379 48.1 2e-05
CE21092 48.1 2e-05
CE26060 48.1 2e-05
Hs13699258 48.1 2e-05
CE03954 48.1 2e-05
CE05752 47.8 2e-05
Hs17455069 47.8 3e-05
CE09969 47.8 3e-05
At2g29150 47.4 3e-05
At2g29370 47.0 4e-05
7293522 47.0 5e-05
CE05726 47.0 5e-05
7304181 46.2 7e-05
YIR036c 46.2 7e-05
At3g47360 46.2 8e-05
CE05526 46.2 8e-05
At2g29260 46.2 9e-05
Hs20070272 45.4 1e-04
CE21093 45.4 1e-04
CE03132 45.1 2e-04
CE15574 45.1 2e-04
At2g29350 45.1 2e-04
YMR226c 45.1 2e-04
SPCC162.03 45.1 2e-04
Hs18582884 44.7 2e-04
CE14538 44.7 2e-04
CE23899 44.7 2e-04
7301617_1 44.7 2e-04
At3g26770 44.7 3e-04
At3g47350 43.9 4e-04
At2g29290 43.9 4e-04
> Hs4502599
Length=277
Score = 160 bits (404), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 89/228 (39%), Positives = 132/228 (57%), Gaps = 9/228 (3%)
Query 45 VFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLD 104
V +VTGGNKGIG + LCR G+ VV+T+RD G A+ +L AEGL LD
Sbjct 7 VALVTGGNKGIGLAIVRDLCRLFSGD---VVLTARDVTRGQAAVQQLQAEGLSPRFHQLD 63
Query 105 ITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITL 164
I +S + ++ +YG +D LVNNAG AFK A P +QA+VT N++ TRD+
Sbjct 64 IDDLQSIRALRDFLRKEYGGLDVLVNNAGIAFKVADPTPFHIQAEVTMKTNFFGTRDVCT 123
Query 165 DMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEKG- 223
+++ L KP R+VNV+S AL+ S EL+ + S++ +E++ +++ F+ +KG
Sbjct 124 ELLPLIKPQGRVVNVSSIMSVRALKSCSPELQQKFRSETITEEELVGLMNKFVEDTKKGV 183
Query 224 -QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
Q+EGWPSS YG++K V L+ ARK S + D ++ CC
Sbjct 184 HQKEGWPSSAYGVTKIGVTVLSRIHARKL----SEQRKGDKILLNACC 227
> Hs4502601
Length=277
Score = 144 bits (364), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 84/240 (35%), Positives = 135/240 (56%), Gaps = 8/240 (3%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
RV +VTG N+GIG A++LCR G+ VV+T+RD G A+ +L AEGL L
Sbjct 6 RVALVTGANRGIGLAIARELCRQFSGD---VVLTARDVARGQAAVQQLQAEGLSPRFHQL 62
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDIT 163
DI +S + ++ +YG ++ LVNNA AFK P ++A++T N++ATR++
Sbjct 63 DIDDLQSIRALRDFLRKEYGGLNVLVNNAAVAFKSDDPMPFDIKAEMTLKTNFFATRNMC 122
Query 164 LDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFI--VACE 221
+++ + KP R+VN++S A + S +L+ R S++ + D+ ++ F+ E
Sbjct 123 NELLPIMKPHGRVVNISSLQCLRAFENCSEDLQERFHSETLTEGDLVDLMKKFVEDTKNE 182
Query 222 KGQQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCCCCD-DGEGKDA 280
++EGWP+S YG+SK V L+ AR+ D +A R +V CC D +GKD+
Sbjct 183 VHEREGWPNSPYGVSKLGVTVLSRILARRLDE--KRKADRILVNACCPGPVKTDMDGKDS 240
> At5g51030
Length=314
Score = 122 bits (306), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 85/241 (35%), Positives = 119/241 (49%), Gaps = 21/241 (8%)
Query 42 AQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEME 101
++ V +VTG N+GIGFE + R L G V++TSRD+ GV+A L G +
Sbjct 36 SETVAVVTGANRGIGFE----MVRQLAGHGLTVILTSRDENVGVEAAKILQEGGFNVDFH 91
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRD 161
LDI S + F IK KYG +D L+NNAG + + V + + NYY T++
Sbjct 92 RLDILDSSSIQEFCEWIKEKYGFIDVLINNAGVNYNVGSDNSVEF-SHMVISTNYYGTKN 150
Query 162 ITLDMMGLFK---PGSRIVNVASAAGEMALQEMSAE---LRHRLMS-KSARQEDIDKVVD 214
I M+ L + G+RIVNV S G + + E +R +LM S +E +DK V
Sbjct 151 IINAMIPLMRHACQGARIVNVTSRLGRLKGRHSKLENEDVRAKLMDVDSLTEEIVDKTVS 210
Query 215 DFIVACEKGQQE--GWPSS--TYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
+F+ E+G E GWP S Y +SK AV A T A++ P E I C
Sbjct 211 EFLKQVEEGTWESGGWPHSFTDYSVSKMAVNAYTRVLAKELSERPEGEK-----IYANCF 265
Query 271 C 271
C
Sbjct 266 C 266
> At1g01800
Length=309
Score = 108 bits (271), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 77/223 (34%), Positives = 126/223 (56%), Gaps = 22/223 (9%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL- 102
RV +VTG NKGIGFE +CR L VV+T+RD+ G+ A+ KL E ++ +
Sbjct 35 RVAVVTGSNKGIGFE----ICRQLANNGITVVLTARDENKGLAAVQKLKTENGFSDQAIS 90
Query 103 ---LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQ-AKVT---CGIN 155
LD++ ++ S A +K+++G +D LVNNAG + + Q A++ N
Sbjct 91 FHPLDVSNPDTIASLAAFVKTRFGKLDILVNNAGVGGANVNVDVLKAQIAEIVEECVKTN 150
Query 156 YYATRDITLDMMGLFKP--GSRIVNVASAAGEMALQEMSAELRHRLMS--KSARQEDIDK 211
YY + + M+ L + RIV++AS G+ L+ +S E ++S ++ +E ID+
Sbjct 151 YYGVKRMCEAMIPLLQSSDSPRIVSIASTMGK--LENVSNEWAKGVLSDAENLTEEKIDE 208
Query 212 VVDDFIVACEKG--QQEGWPS--STYGLSKAAVIALTAAWARK 250
V+++++ ++G Q +GWP+ S Y LSKAAVIALT A++
Sbjct 209 VINEYLKDYKEGALQVKGWPTVMSGYILSKAAVIALTRVLAKR 251
> At5g61830
Length=316
Score = 105 bits (262), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 78/223 (34%), Positives = 117/223 (52%), Gaps = 18/223 (8%)
Query 43 QRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKL--AAEGLKAEM 100
+ V +VTG N+GIGFE A++L + G VV+T+R+ G++A+ L EGLK
Sbjct 36 ENVAVVTGSNRGIGFEIARQLA--VHG--LTVVLTARNVNAGLEAVKSLRHQEEGLKVYF 91
Query 101 ELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATR 160
LD+T S F +K +G +D LVNNAG + + V A+ NY T+
Sbjct 92 HQLDVTDSSSIREFGCWLKQTFGGLDILVNNAGVNYNLGSDNTVEF-AETVISTNYQGTK 150
Query 161 DITLDMMGLFKP---GSRIVNVASAAGEMALQE---MSAELRHRLMSKSARQED-IDKVV 213
++T M+ L +P G+R+VNV+S G + + + ELR +L S E+ ID+ V
Sbjct 151 NMTKAMIPLMRPSPHGARVVNVSSRLGRVNGRRNRLANVELRDQLSSPDLLTEELIDRTV 210
Query 214 DDFIVACEKGQQE--GWPS--STYGLSKAAVIALTAAWARKAD 252
FI + G E GWP + Y +SK AV A T A++ +
Sbjct 211 SKFINQVKDGTWESGGWPQTFTDYSMSKLAVNAYTRLMAKELE 253
> At3g59710
Length=302
Score = 89.4 bits (220), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 70/231 (30%), Positives = 110/231 (47%), Gaps = 21/231 (9%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGL-KAEMELLDI 105
+VTG NKGIGF K+L VV+T+R+ ENG QA L G LDI
Sbjct 32 VVTGANKGIGFAVVKRLLE----LGLTVVLTARNAENGSQAAESLRRIGFGNVHFCCLDI 87
Query 106 TKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITLD 165
+ S +F + G +D LVNNA +F A E + + + N+Y + +T
Sbjct 88 SDPSSIAAFASWFGRNLGILDILVNNAAVSF-NAVGENLIKEPETIIKTNFYGAKLLTEA 146
Query 166 MMGLFKPG---SRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEK 222
++ LF+ SRI+N++S G + + S +R L S+ E ID + F+ +
Sbjct 147 LLPLFRRSVSVSRILNMSSRLGTLN-KLRSPSIRRILESEDLTNEQIDATLTQFLQDVKS 205
Query 223 G--QQEGWPSS--TYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCC 269
G +++GWP + Y +SK A+ A + AR+ D + + + C C
Sbjct 206 GTWEKQGWPENWPDYAISKLALNAYSRVLARRYDG-------KKLSVNCLC 249
> At3g61220
Length=296
Score = 83.6 bits (205), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 73/243 (30%), Positives = 119/243 (48%), Gaps = 48/243 (19%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL- 102
R +VTG N+GIGFE +CR L E VV+TSRD+ G++A+ E LK E+E+
Sbjct 7 RYAVVTGANRGIGFE----ICRQLASEGIRVVLTSRDENRGLEAV-----ETLKKELEIS 57
Query 103 --------LDITKKESRESFVAAIKSKYGHVDSLVNNA-------------------GFA 135
LD+ S S +K+++G +D LVNNA GF
Sbjct 58 DQSLLFHQLDVADPASITSLAEFVKTQFGKLDILVNNAGIGGIITDAEALRAGAGKEGFK 117
Query 136 FKKAATEPVAVQAKVTCGINYYATRDITLDMMGLFK--PGSRIVNVASAAGEMALQEMSA 193
+ + TE + + INYY + + + L K RIVNV+S+ G+ L+ +
Sbjct 118 WDEIITETYELTEEC-IKINYYGPKRMCEAFIPLLKLSDSPRIVNVSSSMGQ--LKNVLN 174
Query 194 ELRHRLMS--KSARQEDIDKVVDDFIVACEKG--QQEGWPS--STYGLSKAAVIALTAAW 247
E ++S ++ +E ID+V++ + ++G +++ W S Y +SKA++ T
Sbjct 175 EWAKGILSDAENLTEERIDQVINQLLNDFKEGTVKEKNWAKFMSAYVVSKASLNGYTRVL 234
Query 248 ARK 250
A+K
Sbjct 235 AKK 237
> At2g24190
Length=296
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 72/259 (27%), Positives = 121/259 (46%), Gaps = 48/259 (18%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL- 102
R IVTGGN+GIGFE +CR L + V++TSRD++ G++A+ E LK E+E+
Sbjct 7 RYAIVTGGNRGIGFE----ICRQLANKGIRVILTSRDEKQGLEAV-----ETLKKELEIS 57
Query 103 --------LDITKKESRESFVAAIKSKYGHVDSLVNNA-------------------GFA 135
LD++ S S +K+ +G +D L+NNA GF
Sbjct 58 DQSIVFHQLDVSDPVSVTSLAEFVKTHFGKLDILINNAGVGGVITDVDALRAGTGKEGFK 117
Query 136 FKKAATEPVAVQAKVTCGINYYATRDITLDMMGLFK--PGSRIVNVASAAGEMALQEMSA 193
+++ TE + A+ INYY + + + L + RI+NV+S G+ ++ +
Sbjct 118 WEETITETYEL-AEECIKINYYGPKRMCEAFIPLLQLSDSPRIINVSSFMGQ--VKNLVN 174
Query 194 ELRHRLMSKSARQED--IDKVVDDFI--VACEKGQQEGWPS--STYGLSKAAVIALTAAW 247
E ++S + + ID+V++ + + + + + W S Y +SKA + A T
Sbjct 175 EWAKGILSDAENLTEVRIDQVINQLLNDLKEDTAKTKYWAKVMSAYVVSKAGLNAYTRIL 234
Query 248 ARKADHCPSMEACRDMVIT 266
A+K C V T
Sbjct 235 AKKHPEIRVNSVCPGFVKT 253
> Hs5031737
Length=280
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 48/125 (38%), Positives = 64/125 (51%), Gaps = 9/125 (7%)
Query 14 LYSIFRSFICRFSPLPRF-LRSKFTFC----TMAQRVFIVTGGNKGIGFETAKKLCRDLK 68
L ++ R + F P R +R T +A RV +VTG GIGF A++L RD
Sbjct 2 LSAVARGYQGWFHPCARLSVRMSSTGIDRKGVLANRVAVVTGSTSGIGFAIARRLARD-- 59
Query 69 GENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSL 128
A VVI+SR ++N +A+AKL EGL + + K E RE VA G VD L
Sbjct 60 --GAHVVISSRKQQNVDRAMAKLQGEGLSVAGIVCHVGKAEDREQLVAKALEHCGGVDFL 117
Query 129 VNNAG 133
V +AG
Sbjct 118 VCSAG 122
> Hs10337605
Length=260
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 52/92 (56%), Gaps = 4/92 (4%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEM 100
+A +V +VT GIGF A++L +D A VV++SR ++N QA+A L EGL
Sbjct 12 LANKVALVTASTDGIGFAIARRLAQD----GAHVVVSSRKQQNVDQAVATLQGEGLSVTG 67
Query 101 ELLDITKKESRESFVAAIKSKYGHVDSLVNNA 132
+ + K E RE VA +G +D LV+NA
Sbjct 68 TVCHVGKAEDRERLVAMAVKLHGGIDILVSNA 99
> At4g09750
Length=322
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 76/155 (49%), Gaps = 13/155 (8%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAE--GLKAEMELLD 104
+VTG N GIGF A+ L A V + R+KE G +AL+K+ +E+ D
Sbjct 47 VVTGANSGIGFAAAEGLAS----RGATVYMVCRNKERGQEALSKIQTSTGNQNVYLEVCD 102
Query 105 ITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITL 164
++ +SF ++ SK V LVNNAG K T P + ++ +N T +T
Sbjct 103 LSSVNEIKSFASSFASKDVPVHVLVNNAGLLENKRTTTPEGFE--LSFAVNVLGTYTMTE 160
Query 165 DMMGLFK---PGSRIVNVASAAGEMALQEMSAELR 196
M+ L + P ++++ VAS G M ++ +L+
Sbjct 161 LMLPLLEKATPDAKVITVAS--GGMYTSPLTTDLQ 193
> 7297326
Length=409
Score = 56.6 bits (135), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 46/171 (26%), Positives = 78/171 (45%), Gaps = 22/171 (12%)
Query 32 LRSKFTFCTMAQRVF----------IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDK 81
+R +F C+ A +V ++TG N GIG+ETA R L ++ R++
Sbjct 100 VRQRFDSCSTALQVLHGKDLHGRTALITGANCGIGYETA----RSLAHHGCEIIFACRNR 155
Query 82 ENGVQALAKLAAEGLKAE----MELLDITKKESRESFVAAIKSKYGHVDSLVNNAG-FAF 136
+ A+ ++A E A LD++ S + FV IK H+D L+ NAG FA
Sbjct 156 SSAEAAIERIAQERPAARSRCRFAALDLSSLRSVQRFVEEIKQSVSHIDYLILNAGVFAL 215
Query 137 KKAATEPVAVQAKVTCGINYYATRDITLDMMGLFKPGSRIVNVASAAGEMA 187
T + T +++ + +TL + LF +RI+ ++S + A
Sbjct 216 PYTRT---VDGLETTFQVSHLSHFYLTLQLETLFDYKTRIIVLSSESHRFA 263
> 7302393
Length=355
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 52/107 (48%), Gaps = 7/107 (6%)
Query 33 RSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDK---ENGVQALA 89
R + R+ ++TGGN+GIG +KL + VV+ RD E V ++
Sbjct 57 RDRVALYKQPDRIAVITGGNRGIGLRIVEKLL----ACDMTVVMGVRDPKIAETAVASIV 112
Query 90 KLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAF 136
L A K E LD+ +S ++F IK +Y VD L+NNAG F
Sbjct 113 DLNATKGKLICEQLDVGDLKSVKAFAQLIKERYSKVDLLLNNAGIMF 159
> At4g05530
Length=254
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 75/161 (46%), Gaps = 20/161 (12%)
Query 28 LPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQA 87
LPR L K V IVT +GIGF ++ E A VV++SR + N +A
Sbjct 5 LPRRLEGK---------VAIVTASTQGIGFGITERFGL----EGASVVVSSRKQANVDEA 51
Query 88 LAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQ 147
+AKL ++G+ A + ++ + R + V KYG +D +V NA +T+P+
Sbjct 52 VAKLKSKGIDAYGIVCHVSNAQHRRNLVEKTVQKYGKIDIVVCNAA---ANPSTDPILSS 108
Query 148 AKVTCG----INYYATRDITLDMMGLFKPGSRIVNVASAAG 184
+ IN ++ + DM + GS ++ + S AG
Sbjct 109 KEAVLDKLWEINVKSSILLLQDMAPHLEKGSSVIFITSIAG 149
> 7300387
Length=317
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 48/152 (31%), Positives = 74/152 (48%), Gaps = 13/152 (8%)
Query 36 FTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKE--NGVQALAKLAA 93
FT M + I+TG N GIG ETAK DL G A +++ R+ E N V+
Sbjct 26 FTETKMEGKTVIITGANSGIGKETAK----DLAGRGARIIMACRNLETANAVKDEIVKET 81
Query 94 EGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAG--FAFKKAATEPVAVQAKVT 151
+ K ++ LD+ ++S F A I +D L++NAG AF+ +E ++T
Sbjct 82 KNNKILVKKLDLGSQKSVREFAADIVKTEPKIDVLIHNAGMALAFRGQTSED---GVELT 138
Query 152 CGINYYATRDITLDMMGLFKPG--SRIVNVAS 181
N+Y +T ++ + K +RIV VAS
Sbjct 139 MATNHYGPFLLTHLLIDVLKKSAPARIVIVAS 170
> 7290108
Length=270
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 73/146 (50%), Gaps = 9/146 (6%)
Query 40 TMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE 99
+++ +V IVTG + GIG A+ L R E A + + R+ N ++A K + +G +AE
Sbjct 2 SLSNKVVIVTGASSGIGAAIAQVLAR----EGATLALVGRNVAN-LEATKK-SLKGTQAE 55
Query 100 MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYAT 159
+ + D+TK ++ V +K+G +D LVNNAG K + + N
Sbjct 56 IVVADVTKDA--DAIVQQTLAKFGRIDVLVNNAGILGKGGLIDLDIEEFDAVLNTNLRGV 113
Query 160 RDITLDMM-GLFKPGSRIVNVASAAG 184
+T ++ L K +VNV+S AG
Sbjct 114 ILLTKAVLPHLLKTKGAVVNVSSCAG 139
> Hs21553325
Length=330
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 46/98 (46%), Gaps = 6/98 (6%)
Query 38 FCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLK 97
F RV IVTGG GIG+ TAK L R V+I + Q ++K+ E L
Sbjct 38 FPPRPDRVAIVTGGTDGIGYSTAKHLAR----LGMHVIIAGNNDSKAKQVVSKIKEETLN 93
Query 98 AEMELL--DITKKESRESFVAAIKSKYGHVDSLVNNAG 133
++E L D+ S FV K K + L+NNAG
Sbjct 94 DKVEFLYCDLASMTSIRQFVQKFKMKKIPLHVLINNAG 131
> 7295487
Length=317
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 51/168 (30%), Positives = 78/168 (46%), Gaps = 23/168 (13%)
Query 37 TFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGL 96
T +A +V +VT GIGF AK+L D A VVI+SR ++N ALA+L L
Sbjct 65 TMKRLAGKVAVVTASTDGIGFAIAKRLAED----GAAVVISSRKQKNVDSALAELRKLNL 120
Query 97 KAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTC---- 152
+++ E R+ SK+G ++ LV+N AAT P AV + C
Sbjct 121 NVHGLKCHVSEPEDRKQLFEETISKFGKLNILVSN-------AATNP-AVGGVLECDEKV 172
Query 153 -----GINYYATRDITLDMMGLFKP--GSRIVNVASAAGEMALQEMSA 193
+N ++ + + + L + S IV V+S AG A + + A
Sbjct 173 WDKIFDVNVKSSYLLAKEALPLLRQQKNSSIVFVSSIAGYDAFELLGA 220
> Hs7706523
Length=414
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 55/102 (53%), Gaps = 7/102 (6%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEME-- 101
+V +VTG N GIGFETAK L G A V++ R+ +A++++ E KA++E
Sbjct 125 KVVVVTGANSGIGFETAKSFA--LHG--AHVILACRNMARASEAVSRILEEWHKAKVEAM 180
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLV-NNAGFAFKKAATE 142
LD+ S + F A K+K + LV N A FA + T+
Sbjct 181 TLDLALLRSVQHFAEAFKAKNVPLHVLVCNAATFALPWSLTK 222
> 7300379
Length=266
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 52/114 (45%), Gaps = 22/114 (19%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAE-GLK-- 97
+ +++ +VTGGN GIGFE A+ L G +++ R+ E G +A A + E G +
Sbjct 41 IKEQIVVVTGGNSGIGFEIAQALA----GRGGRIILACRNLEAGKRAAAIIKRELGCRTP 96
Query 98 ---------------AEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAF 136
E LD+ S F + +++ +D LVNNAG F
Sbjct 97 LNSLDEDDNPEDRYFVEARYLDLCSLRSVHHFAGQLMAEFERIDVLVNNAGVVF 150
> CE21092
Length=278
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 53/98 (54%), Gaps = 7/98 (7%)
Query 42 AQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE-- 99
+ +V I+TG + GIG TA L + E A V +T R E + + ++ G ++
Sbjct 5 SDKVAIITGSSSGIGRSTAVLLAQ----EGAKVTVTGRSSEKIQETVNEIHKNGGSSDNI 60
Query 100 -MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAF 136
+ L D+ + E ++ + + S++G +D L+NNAG AF
Sbjct 61 NIVLGDLNESECQDELIKSTLSRFGKIDILINNAGAAF 98
> CE26060
Length=347
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 59/192 (30%), Positives = 90/192 (46%), Gaps = 22/192 (11%)
Query 12 FSLYSIFRSFICRFSPLPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGEN 71
F L+ I FI F +P R F F +R IVTGG+KGIGF+ A L +G +
Sbjct 14 FILFLIVLLFI--FFSIPS--RRDFHFF---RRHAIVTGGSKGIGFQLAVGLIE--RGCH 64
Query 72 AVVVITS-RDKENG---VQALAKLAAEGLKAEMELLDITKKES--RESFVAAIKSKYGHV 125
+V + +D E +Q LA + K + +D+T + +F A + + G +
Sbjct 65 VTIVARNVKDLEKACADLQVLADQRGQRQKVHWKSIDMTGGYDVIKSAFDDAAR-ELGPI 123
Query 126 DSLVNNAGFAFKKAATEPVAVQAKVTCGINY----YATRDITLDMMGLFKPGSRIVNVAS 181
D L+NNAG + + E + +NY +ATR + DM + I V+S
Sbjct 124 DILINNAGHSVQAPFCELPITDFEKQMAVNYLSAVHATRAVVDDMKT--RKTGHISFVSS 181
Query 182 AAGEMALQEMSA 193
AAG+ A+ SA
Sbjct 182 AAGQFAIFGYSA 193
> Hs13699258
Length=363
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 55/102 (53%), Gaps = 7/102 (6%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEME-- 101
+V +VTG N GIGFETAK L G A V++ R+ +A++++ E KA++E
Sbjct 125 KVVVVTGANSGIGFETAKSFA--LHG--AHVILACRNMARASEAVSRILEEWHKAKVETM 180
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLV-NNAGFAFKKAATE 142
LD+ S + F A K+K + LV N A FA + T+
Sbjct 181 TLDLALLRSVQHFAEAFKAKNVPLHVLVCNAATFALPWSLTK 222
> CE03954
Length=274
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 46/93 (49%), Gaps = 7/93 (7%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDK---ENGVQALAKLAAEGLKAEM 100
+V I+TG + GIG TA L D A V IT RD E QA+ K
Sbjct 7 KVAIITGSSNGIGRATAVLLATD----GAKVTITGRDAARLEETRQAILKAGISATNVNS 62
Query 101 ELLDITKKESRESFVAAIKSKYGHVDSLVNNAG 133
+ D+T E ++ +++ K+G ++ L+NNAG
Sbjct 63 VVADVTTAEGQDLLISSTLDKFGKINILINNAG 95
> CE05752
Length=284
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 54/109 (49%), Gaps = 12/109 (11%)
Query 34 SKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAA 93
S+FT +V I+TG + GIG TA R L E A V +T R+ E + L
Sbjct 2 SRFT-----DKVAIITGSSNGIGQATA----RLLASEGAKVTVTGRNAERLEETKNILLG 52
Query 94 EGLKAEMELL---DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKA 139
G+ L+ DIT++ +E+ + + K+G +D LVNNAG A
Sbjct 53 AGVPEGNVLVVVGDITQESVQENLIKSTLDKFGKIDILVNNAGAGIPDA 101
> Hs17455069
Length=313
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 74/153 (48%), Gaps = 11/153 (7%)
Query 40 TMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE 99
++++ +TG + G G AK+L + + V+ + E G Q L + + L+
Sbjct 22 NLSEKYVFITGCDSGFGNLLAKQLV-----DRGMQVLAACFTEEGSQKLQRDTSYRLQTT 76
Query 100 MELLDITKKESRESFVAAIKSKYGH--VDSLVNNAGFAFKKAATEPVAVQAKV-TCGINY 156
LLD+TK ES ++ ++ K G + +LVNNAG E + V +N
Sbjct 77 --LLDVTKSESIKAAAQWVRDKVGEQGLWALVNNAGVGLPSGPNEWLTKDDFVKVINVNL 134
Query 157 YATRDITLDMMGLFKPG-SRIVNVASAAGEMAL 188
++TL M+ + K R+VN++S+ G +A+
Sbjct 135 VGLIEVTLHMLPMVKRARGRVVNMSSSGGRVAV 167
> CE09969
Length=257
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 49/107 (45%), Gaps = 13/107 (12%)
Query 26 SPLPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGV 85
SP +FL RV +VT KGIGF AK+L A VV+ SR KEN
Sbjct 3 SPATKFL---------TDRVALVTASTKGIGFAIAKQLG----AAGASVVVCSRKKENVD 49
Query 86 QALAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNA 132
+A+A L E + A + K R + ++ +D LV+NA
Sbjct 50 EAVAALRLENIDAHGTTAHVGNKSDRTKLIDFTLDRFTKLDILVSNA 96
> At2g29150
Length=268
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 53/100 (53%), Gaps = 5/100 (5%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDIT 106
+VTGG+KG+G + + +L A V +RD+ + L + A+G + + D++
Sbjct 22 LVTGGSKGLG----EAVVEELAMLGARVHTCARDETQLQERLREWQAKGFEVTTSVCDVS 77
Query 107 KKESRESFVAAIKSKY-GHVDSLVNNAGFAFKKAATEPVA 145
+E RE + + S + G ++ LVNNAG K +TE A
Sbjct 78 SREQREKLMETVSSVFQGKLNILVNNAGTGIIKPSTEYTA 117
> At2g29370
Length=268
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 70/149 (46%), Gaps = 7/149 (4%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDIT 106
+VTGG+KG+G K + +L A V +RD+ ++L + A+GL+ + D++
Sbjct 22 LVTGGSKGLG----KAVVEELAMLGARVHTCARDETQLQESLREWQAKGLQVTTSVCDVS 77
Query 107 KKESRESFVAAIKSKY-GHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITLD 165
++ RE + + S + G + LV N G K TE A + N +T +
Sbjct 78 SRDQREKLMETVSSLFQGKLSILVPNVGIGVLKPTTECTAEEFSFIIATNLESTFHFSQL 137
Query 166 MMGLFK-PGS-RIVNVASAAGEMALQEMS 192
L K GS IV ++S AG + L S
Sbjct 138 AHPLLKASGSGNIVLMSSVAGVVNLGNTS 166
> 7293522
Length=242
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 43/147 (29%), Positives = 69/147 (46%), Gaps = 13/147 (8%)
Query 38 FCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLK 97
+ +A +V +VTG GIG + L + L A V+ +R E +Q L +A + +
Sbjct 2 WTDLAGKVILVTGAGAGIG----QALVKQLASAGATVIAVARKPEQ-LQQL--VAFDPVH 54
Query 98 AEMELLDITKKES-RESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINY 156
+ LD++ ++ RE +K +D LVNNAG A K E +N
Sbjct 55 IQPLQLDLSGWQAVREGL-----AKVPLLDGLVNNAGVAIIKPFEELTEQDFDTHFDVNI 109
Query 157 YATRDITLDMMGLFKPGSRIVNVASAA 183
A ++T ++ K G+ IVNV+S A
Sbjct 110 KAVFNVTQSLLPRLKDGASIVNVSSIA 136
> CE05726
Length=277
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 45/142 (31%), Positives = 67/142 (47%), Gaps = 12/142 (8%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDK---ENGVQALAKLAAEGLKAEMELL 103
I+TG + GIG A + E A V IT R++ E Q + K K +
Sbjct 10 IITGSSNGIGRSAAVIFAK----EGAQVTITGRNEDRLEETKQQILKAGVPAEKINAVVA 65
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAG--FAFKKAAT-EPVAVQAKVTCGINYYATR 160
D+T+ ++ + +K+G +D LVNNAG A A T +PV + K T +N+ A
Sbjct 66 DVTEASGQDDIINTTLAKFGKIDILVNNAGANLADGTANTDQPVELYQK-TFKLNFQAVI 124
Query 161 DITLDMM-GLFKPGSRIVNVAS 181
++T L K IVNV+S
Sbjct 125 EMTQKTKEHLIKTKGEIVNVSS 146
> 7304181
Length=331
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 47/160 (29%), Positives = 79/160 (49%), Gaps = 17/160 (10%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEM--E 101
+VFIVTG N GIG ET R++ V + R+ + +A ++ E +
Sbjct 46 KVFIVTGANTGIGKET----VREIAKRGGTVYMACRNLKKCEEAREEIVLETKNKYVYCR 101
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAF--KKAATEPVAVQAKVTCGINYYAT 159
D+ +ES FVAA K + H+ L+NNAG + ++ + +Q G+N+
Sbjct 102 QCDLASQESIRHFVAAFKREQEHLHVLINNAGVMRCPRSLTSDGIELQ----LGVNHMGH 157
Query 160 RDITLDMMGLFKPG--SRIVNVASAA---GEMALQEMSAE 194
+T ++ L K SRIVNV+S A GE+ +++++
Sbjct 158 FLLTNLLLDLLKKSSPSRIVNVSSLAHTRGEINTGDLNSD 197
> YIR036c
Length=263
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 48/91 (52%), Gaps = 4/91 (4%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
+V ++TG ++GIG + K + + ++ +V E G+Q+L + K +L
Sbjct 3 KVILITGASRGIGLQLVKTVIEE---DDECIVYGVARTEAGLQSLQREYGAD-KFVYRVL 58
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAGF 134
DIT + E+ V I+ K+G +D +V NAG
Sbjct 59 DITDRSRMEALVEEIRQKHGKLDGIVANAGM 89
> At3g47360
Length=309
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 48/188 (25%), Positives = 82/188 (43%), Gaps = 18/188 (9%)
Query 4 FLWSFNSKFSLYSIFRSFICRFSPLPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKL 63
FL+ F + Y + + +C LR F +A++V ++TG + GIG A +
Sbjct 19 FLFLF---YPFYLLIKLVLC--------LRKNLHFENVARKVVLITGASSGIGEHVAYEY 67
Query 64 CRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYG 123
+ KG +V RD+ V ++ G + D++ E + F+ +G
Sbjct 68 AK--KGAYLALVARRRDRLEIVAETSRQLGSG-NVIIIPGDVSNVEDCKKFIDETIRHFG 124
Query 124 HVDSLVNNAGFAFKKAATEPVA-VQ-AKVTCGINYYATRDIT-LDMMGLFKPGSRIVNVA 180
+D L+NNAG F+ E +Q A IN++ T IT + L K +IV +
Sbjct 125 KLDHLINNAG-VFQTVLFEDFTQIQDANPIMDINFWGTTYITYFAIPHLRKSKGKIVAIT 183
Query 181 SAAGEMAL 188
S + + L
Sbjct 184 SGSANIPL 191
> CE05526
Length=278
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 49/98 (50%), Gaps = 7/98 (7%)
Query 42 AQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE-- 99
A++V I+TG + GIG TA R E A V IT R E + ++ A G+ +
Sbjct 5 AEKVAIITGSSNGIGRATAVLFAR----EGAKVTITGRHAERLEETRQQILAAGVSEQNV 60
Query 100 -MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAF 136
+ D+T ++ ++ K+G +D LVNNAG A
Sbjct 61 NSVVADVTTDAGQDEILSTTLGKFGKLDILVNNAGAAI 98
> At2g29260
Length=322
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 37/152 (24%), Positives = 71/152 (46%), Gaps = 9/152 (5%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDIT 106
+VTGG +GIG + + +L G A V +R++ L+ G + + D++
Sbjct 74 LVTGGTRGIG----RAIVEELAGLGAEVHTCARNEYELENCLSDWNRSGFRVAGSVCDVS 129
Query 107 KKESRESFVAAIKSKY-GHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINY---YATRDI 162
+ RE+ + + S + G + LVNN G +K E A + N+ + +
Sbjct 130 DRSQREALMETVSSVFEGKLHILVNNVGTNIRKPMVEFTAGEFSTLMSTNFESVFHLCQL 189
Query 163 TLDMMGLFKPGSRIVNVASAAGEMALQEMSAE 194
++ K GS +V ++S +G ++L+ MS +
Sbjct 190 AYPLLRESKAGS-VVFISSVSGFVSLKNMSVQ 220
> Hs20070272
Length=318
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/148 (28%), Positives = 68/148 (45%), Gaps = 10/148 (6%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENG--VQALAKLAAEGLKAEME 101
+V +VTG N GIG ETAK+L + A V + RD E G V + + +
Sbjct 42 KVVVVTGANTGIGKETAKELAQ----RGARVYLACRDVEKGELVAKEIQTTTGNQQVLVR 97
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRD 161
LD++ +S +F ++ H+ L+NNAG + + + G+N+
Sbjct 98 KLDLSDTKSIRAFAKGFLAEEKHLHVLINNAGVMMCPYSKTADGFEMHI--GVNHLGHFL 155
Query 162 ITLDMMGLFKPG--SRIVNVASAAGEMA 187
+T ++ K SRIVNV+S A +
Sbjct 156 LTHLLLEKLKESAPSRIVNVSSLAHHLG 183
> CE21093
Length=281
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 50/170 (29%), Positives = 77/170 (45%), Gaps = 17/170 (10%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL- 102
+V +VTG + GIG A +D A V +T R+ E + ++ G+ L
Sbjct 8 KVALVTGSSNGIGRAAAVLFAKD----GAKVTVTGRNAERLEETRQEILKSGVPESHVLS 63
Query 103 --LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAF-----KKAATEPVAVQAKVTCGIN 155
D+ ++ ++ V + K+G +D LVNNAG AF + + V+V K+ IN
Sbjct 64 VATDLAAEKGQDELVNSTIQKFGRLDILVNNAGAAFNDDQGRVGVDQDVSVYDKIMQ-IN 122
Query 156 YYATRDITLDMM-GLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSA 204
+ +T L K IVNV+S AG Q + + MSKSA
Sbjct 123 MRSVVTLTQKAKEHLVKAKGEIVNVSSIAGTAHAQP---GVMYYAMSKSA 169
> CE03132
Length=278
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 51/101 (50%), Gaps = 7/101 (6%)
Query 42 AQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEME 101
+++V I+TG + GIG ETA + E A V +T R +E + L G+K
Sbjct 6 SKKVAIITGSSSGIGRETALLFAK----EGAKVTVTGRSEEKLEETKKALLDAGIKESNF 61
Query 102 LL---DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKA 139
L+ DIT ++ ++ K+G ++ LVNNAG + A
Sbjct 62 LIVPADITFSTGQDELISQTLKKFGRINILVNNAGASIPDA 102
> CE15574
Length=279
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 55/186 (29%), Positives = 79/186 (42%), Gaps = 27/186 (14%)
Query 28 LPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQA 87
+PRF + +V +VTG + GIG TA L R E A V IT R+ + +
Sbjct 1 MPRF----------SGKVALVTGSSNGIGRATAILLAR----EGAKVTITGRNAQRLEET 46
Query 88 LAKLAAEGLKAEMEL---LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPV 144
++ G+ + L D+ + + + + +G +D LVNNAG A +
Sbjct 47 KQEILRSGVPEDHVLSIIADLATESGQIELMNSTVDIFGRLDILVNNAGAAITDLEGH-I 105
Query 145 AVQAKV-----TCGINYYATRDITLDMM-GLFKPGSRIVNVASAAGEMALQEMSAELRHR 198
V V T IN + +T L K IVNV+S AG Q EL +
Sbjct 106 GVGTNVSVFDKTMRINLRSVVTLTQKAKEHLIKTKGEIVNVSSIAGG---QHAQPELIYY 162
Query 199 LMSKSA 204
MSKSA
Sbjct 163 AMSKSA 168
> At2g29350
Length=269
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 67/141 (47%), Gaps = 7/141 (4%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDIT 106
+VTGG+KGIG + + +L A V +RD+ + L + A+G + + D++
Sbjct 21 LVTGGSKGIG----EAVVEELAMLGAKVHTCARDETQLQERLREWQAKGFQVTTSVCDVS 76
Query 107 KKESRESFVAAIKSKY-GHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITLD 165
++ R + + S Y G ++ LVNN G + K TE A N + ++
Sbjct 77 SRDQRVKLMETVSSLYQGKLNILVNNVGTSIFKPTTEYTAEDFSFVMATNLESAFHLSQL 136
Query 166 MMGLFK-PGS-RIVNVASAAG 184
L K GS IV ++SAAG
Sbjct 137 AHPLLKASGSGSIVLISSAAG 157
> YMR226c
Length=267
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/148 (26%), Positives = 69/148 (46%), Gaps = 4/148 (2%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSR-DKENGVQALAKLAAEGLKAE 99
+A++ ++TG + GIG TA + G+ +++ R +K ++ K
Sbjct 11 LAKKTVLITGASAGIGKATALEYLEASNGDMKLILAARRLEKLEELKKTIDQEFPNAKVH 70
Query 100 MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQ-AKVTCGINYYA 158
+ LDIT+ E + F+ + ++ +D LVNNAG A +A + + N A
Sbjct 71 VAQLDITQAEKIKPFIENLPQEFKDIDILVNNAGKALGSDRVGQIATEDIQDVFDTNVTA 130
Query 159 TRDITLDMMGLF--KPGSRIVNVASAAG 184
+IT ++ +F K IVN+ S AG
Sbjct 131 LINITQAVLPIFQAKNSGDIVNLGSIAG 158
> SPCC162.03
Length=292
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 46/157 (29%), Positives = 76/157 (48%), Gaps = 27/157 (17%)
Query 35 KFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAE 94
K TF T+ ++TG +KG+G+ K +G N V+ SR + +KL
Sbjct 2 KSTFSTV-----LITGSSKGLGYALVK--VGLAQGYN--VIACSRAPDTITIEHSKL--- 49
Query 95 GLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFA----FKKAATEPVAVQAKV 150
LK + LD+T +S E+ K ++G+VD ++NNAG+ F+ E + Q V
Sbjct 50 -LKLK---LDVTDVKSVETAFKDAKRRFGNVDIVINNAGYGLVGEFESYNIEEMHRQMNV 105
Query 151 TCGINYYATRDITLDMMGLFKP---GSRIVNVASAAG 184
N++ IT + + L + G RI+ ++S AG
Sbjct 106 ----NFWGVAYITKEALNLMRESGKGGRILQISSVAG 138
> Hs18582884
Length=316
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 50/200 (25%), Positives = 91/200 (45%), Gaps = 24/200 (12%)
Query 12 FSLYSIFRSFICRFSPLPRFLRSKF-------TFCTMAQRVFIVTGGNKGIGFETAKKLC 64
L + F SF+ +P R KF T + +V ++TG N GIG ETA
Sbjct 5 LGLLTSFFSFLYMVAPSIR----KFFAGGVCRTNVQLPGKVVVITGANTGIGKETA---- 56
Query 65 RDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL--LDITKKESRESFVAAIKSKY 122
R+L A V I RD G A +++ + +++ + LD++ +S +F ++
Sbjct 57 RELASRGARVYIACRDVLKGESAASEIRVDTKNSQVLVRKLDLSDTKSIRAFAEGFLAEE 116
Query 123 GHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITLDMMGLFKPG--SRIVNVA 180
+ L+NNAG + + + G+N+ +T ++ K +R+VNV+
Sbjct 117 KQLHILINNAGVMMCPYSKTADGFETHL--GVNHLGHFLLTYLLLERLKVSAPARVVNVS 174
Query 181 SAA---GEMALQEMSAELRH 197
S A G++ ++ +E R+
Sbjct 175 SVAHHIGKIPFHDLQSEKRY 194
> CE14538
Length=280
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 48/100 (48%), Gaps = 11/100 (11%)
Query 41 MAQR----VFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKE---NGVQALAKLAA 93
MAQR V IVTG + GIG TA L E A V IT R+ E QAL K+
Sbjct 1 MAQRFTDEVAIVTGSSNGIGRATAILLAS----EGAKVTITGRNAERLEESRQALLKVGV 56
Query 94 EGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAG 133
+ D+T ++ + + K+G ++ L+NNAG
Sbjct 57 PSGHINSVVADVTTGAGQDVLIDSTLKKFGKINILINNAG 96
> CE23899
Length=279
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 50/97 (51%), Gaps = 7/97 (7%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL- 102
+V +VTG + GIG TA L + E A V IT R+ + + ++ G+ + L
Sbjct 7 KVALVTGSSNGIGRATAVLLAQ----EGAKVTITGRNADRLEETRQEILKSGVPEDHVLS 62
Query 103 --LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFK 137
D+ ++ ++ V + K+G +D LVNNAG AF
Sbjct 63 IATDLATEKGQDELVNSTIQKFGRLDILVNNAGAAFN 99
> 7301617_1
Length=302
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/97 (32%), Positives = 49/97 (50%), Gaps = 7/97 (7%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAE-----G 95
+A R +TG ++GIG E A K RD G N VV + + + AAE G
Sbjct 7 LAGRTLFITGASRGIGKEIALKAARD--GANIVVAAKTAEPHPKLPGTIYSAAEEIEKAG 64
Query 96 LKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNA 132
KA ++D+ ++ S V A +K+G +D ++NNA
Sbjct 65 GKAYPCVVDVRDEQQVRSAVEAAVAKFGGIDIVINNA 101
> At3g26770
Length=280
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 47/101 (46%), Gaps = 7/101 (6%)
Query 34 SKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAA 93
+KF + +V ++TGG G+G TA + R A VVI D E G + +L +
Sbjct 8 TKFYSKKLEGKVALITGGASGLGKATASEFLR----HGARVVIADLDAETGTKTAKELGS 63
Query 94 EGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGF 134
E AE D+T + V +YG +D + NNAG
Sbjct 64 E---AEFVRCDVTVEADIAGAVEMTVERYGKLDVMYNNAGI 101
> At3g47350
Length=308
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 46/191 (24%), Positives = 82/191 (42%), Gaps = 14/191 (7%)
Query 12 FSLYSIFRSFICRFSPLPRFLR-----SKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRD 66
F L + SF+ F P F + F + +V ++TG + GIG A + +
Sbjct 10 FLLPPLTISFLVLFYPFYLFTKLMSCLKHLHFENVTGKVVLITGASSGIGEHVAYEYAK- 68
Query 67 LKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL--DITKKESRESFVAAIKSKYGH 124
KG +V +D+ V ++ G ++ ++ D++ E + F+ +G
Sbjct 69 -KGAKLALVARRKDRLEIVAETSRQLGSG---DVIIIPGDVSNVEDCKKFIDETIHHFGK 124
Query 125 VDSLVNNAGFAFKKAATEPVAVQ-AKVTCGINYYATRDIT-LDMMGLFKPGSRIVNVASA 182
+D L+NNAG + +Q A IN++ + IT + L K +IV ++SA
Sbjct 125 LDHLINNAGVPQTVIFEDFTQIQDANSIMDINFWGSTYITYFAIPHLRKSKGKIVVISSA 184
Query 183 AGEMALQEMSA 193
+ LQ S
Sbjct 185 TAIIPLQAASV 195
> At2g29290
Length=262
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 49/100 (49%), Gaps = 5/100 (5%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDIT 106
+VTGG KGIG + + +L A V +RD+ + L + +G + + D++
Sbjct 13 LVTGGTKGIG----EAVVEELSILGARVHTCARDETQLQERLREWQEKGFQVTTSICDVS 68
Query 107 KKESRESFVAAIKSKY-GHVDSLVNNAGFAFKKAATEPVA 145
+E RE + + S + G ++ LVNN G K TE A
Sbjct 69 LREQREKLMETVSSLFQGKLNILVNNVGTLMLKPTTEYTA 108
Lambda K H
0.322 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6339830234
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40