bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1389_orf1
Length=190
Score E
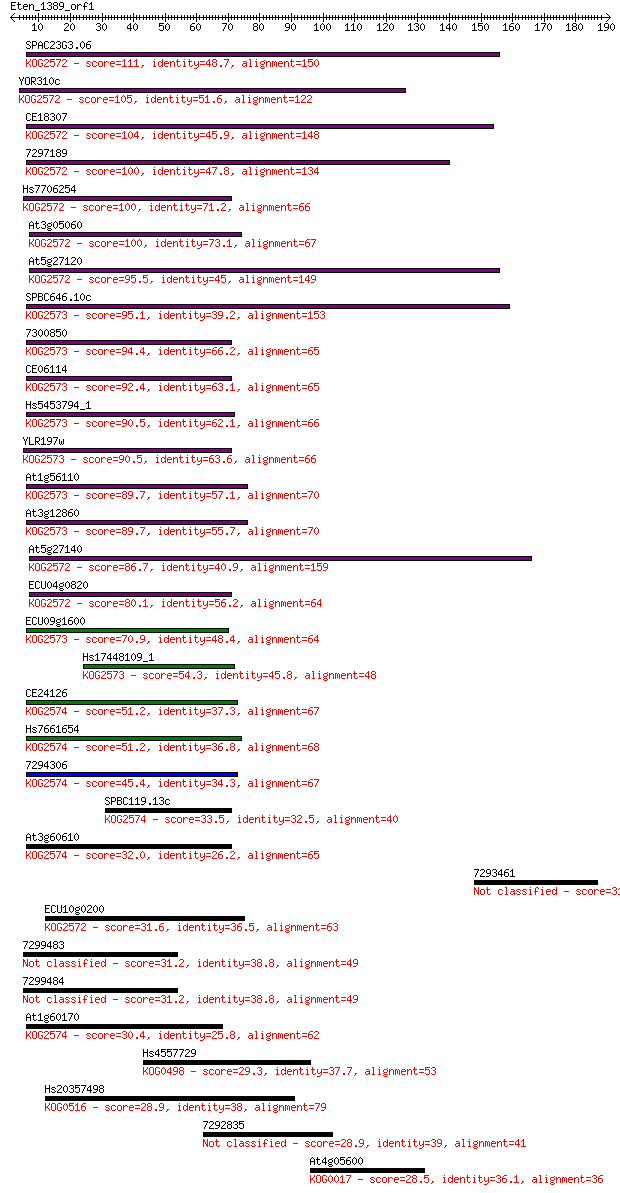
Sequences producing significant alignments: (Bits) Value
SPAC23G3.06 111 1e-24
YOR310c 105 4e-23
CE18307 104 1e-22
7297189 100 2e-21
Hs7706254 100 2e-21
At3g05060 100 2e-21
At5g27120 95.5 6e-20
SPBC646.10c 95.1 7e-20
7300850 94.4 1e-19
CE06114 92.4 5e-19
Hs5453794_1 90.5 2e-18
YLR197w 90.5 2e-18
At1g56110 89.7 3e-18
At3g12860 89.7 3e-18
At5g27140 86.7 3e-17
ECU04g0820 80.1 2e-15
ECU09g1600 70.9 2e-12
Hs17448109_1 54.3 2e-07
CE24126 51.2 1e-06
Hs7661654 51.2 1e-06
7294306 45.4 6e-05
SPBC119.13c 33.5 0.27
At3g60610 32.0 0.73
7293461 31.6 0.94
ECU10g0200 31.6 1.1
7299483 31.2 1.3
7299484 31.2 1.3
At1g60170 30.4 2.0
Hs4557729 29.3 4.6
Hs20357498 28.9 6.8
7292835 28.9 7.0
At4g05600 28.5 8.1
> SPAC23G3.06
Length=508
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 73/150 (48%), Positives = 86/150 (57%), Gaps = 29/150 (19%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
ASTIQILGAEKALFRALKTK +TPKYG+IYHA+LVGQA K KGKI+RVLA K +L +RV
Sbjct 314 ASTIQILGAEKALFRALKTKHSTPKYGLIYHASLVGQANSKNKGKIARVLATKAALSLRV 373
Query 66 DALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAIACRRYVENRLEQLEQQLAGSGPK 125
DAL++ NGN + + R VENRL LE G
Sbjct 374 DALSDK----------DTTNGN------------IGLENRIRVENRLRSLE---GGKLLP 408
Query 126 PPSKPAFQRYEPHRETNGVSKKYDVSTDAV 155
P+ P Q + NG S Y +TDAV
Sbjct 409 LPTAPVQQ---SKVQINGTS-AYSTATDAV 434
> YOR310c
Length=511
Score = 105 bits (263), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 63/122 (51%), Positives = 78/122 (63%), Gaps = 22/122 (18%)
Query 4 SSASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCV 63
S ASTIQILGAEKALFRALKTK +TPKYG++YHA+LVGQAT K KGKI+RVLAAK ++ +
Sbjct 312 SPASTIQILGAEKALFRALKTKHDTPKYGLLYHASLVGQATGKNKGKIARVLAAKAAVSL 371
Query 64 RVDALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAIACRRYVENRLEQLEQQLAGSG 123
R DAL E + +G+ + + R VENRL QLE + +
Sbjct 372 RYDALAEDRD----------DSGD------------IGLESRAKVENRLSQLEGRDLRTT 409
Query 124 PK 125
PK
Sbjct 410 PK 411
> CE18307
Length=487
Score = 104 bits (259), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 68/148 (45%), Positives = 83/148 (56%), Gaps = 28/148 (18%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
ASTIQILGAEKALFRALKTK +TPKYG+IYHA L+ QA PK+KGK++R LAAK SL R+
Sbjct 311 ASTIQILGAEKALFRALKTKKDTPKYGLIYHAQLITQAPPKVKGKMARKLAAKCSLATRI 370
Query 66 DALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAIACRRYVENRLEQLEQQLAGSGPK 125
DAL++ SA N + I CR +EN L + E + S K
Sbjct 371 DALSDE----------SATN-------------EIGIECRAALENVL-RTESERGPSKNK 406
Query 126 PPSKPAFQRYEPHRETNGVSKKYDVSTD 153
+YE ET +YD S D
Sbjct 407 SFGSHRHDKYEFKSET----YEYDASAD 430
> 7297189
Length=454
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 64/138 (46%), Positives = 81/138 (58%), Gaps = 31/138 (22%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
+ST+QILGAEKALFRALKTK +TPKYG+IYHA LVGQA+ K KGK+SR LAAK SL RV
Sbjct 314 SSTVQILGAEKALFRALKTKKDTPKYGLIYHAQLVGQASQKNKGKMSRSLAAKASLATRV 373
Query 66 DALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAIACRRYVENRLEQLEQ----QLAG 121
DA E A AA + +E+RL LE+ +L+G
Sbjct 374 DAFGEEATFELGAAH------------------------KVKLESRLRLLEEGNLRKLSG 409
Query 122 SGPKPPSKPAFQRYEPHR 139
+G +K F++Y+ R
Sbjct 410 TG---KAKAKFEKYQAKR 424
> Hs7706254
Length=529
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/66 (71%), Positives = 57/66 (86%), Gaps = 0/66 (0%)
Query 5 SASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVR 64
+AST+QILGAEKALFRALK++ +TPKYG+IYHA+LVGQ +PK KGKISR+LAAK L +R
Sbjct 312 AASTVQILGAEKALFRALKSRRDTPKYGLIYHASLVGQTSPKHKGKISRMLAAKTVLAIR 371
Query 65 VDALTE 70
DA E
Sbjct 372 YDAFGE 377
> At3g05060
Length=533
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 49/67 (73%), Positives = 57/67 (85%), Gaps = 0/67 (0%)
Query 7 STIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRVD 66
ST+QILGAEKALFRALKTK TPKYG+I+HA+LVGQA PK KGKISR LAAK L +RVD
Sbjct 313 STVQILGAEKALFRALKTKHATPKYGLIFHASLVGQAAPKHKGKISRSLAAKTVLAIRVD 372
Query 67 ALTEAAE 73
AL ++ +
Sbjct 373 ALGDSQD 379
> At5g27120
Length=533
Score = 95.5 bits (236), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 67/158 (42%), Positives = 89/158 (56%), Gaps = 36/158 (22%)
Query 7 STIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRVD 66
ST+QILGAEKALFRALKTK TPKYG+I+HA++VGQA PK KGKISR LAAK L +R D
Sbjct 312 STVQILGAEKALFRALKTKHATPKYGLIFHASVVGQAAPKNKGKISRSLAAKSVLAIRCD 371
Query 67 ALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAIACRRYVENRLEQLE----QQLAGS 122
AL G S N T+ + R +E RL LE +L+GS
Sbjct 372 AL-----------GDSQDN-------------TMGVENRLKLEARLRTLEGKDLGRLSGS 407
Query 123 GPKPPSKPAFQRYEPHRE--TNGV---SKKYDVSTDAV 155
KP + Y+ ++ + G+ +K Y+ + D++
Sbjct 408 A---KGKPKIEVYDKDKKKGSGGLITPAKTYNTAADSL 442
> SPBC646.10c
Length=497
Score = 95.1 bits (235), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 60/155 (38%), Positives = 77/155 (49%), Gaps = 31/155 (20%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
AST+QILGAEKALFRALKT+ NTPKYGIIYH++ +G+A K KG+ISR LA K S+ R+
Sbjct 326 ASTVQILGAEKALFRALKTRGNTPKYGIIYHSSFIGKAGAKNKGRISRFLANKCSIASRI 385
Query 66 DALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAI--ACRRYVENRLEQLEQQLAGSG 123
D ++A PT A RR VE RL + G
Sbjct 386 DNFSDA--------------------------PTTAFGQVLRRQVEERLNFFD---TGVA 416
Query 124 PKPPSKPAFQRYEPHRETNGVSKKYDVSTDAVDTA 158
P S + YE + + +V D +T
Sbjct 417 PTRNSIAMAEAYEKALSSVNIDGDEEVDIDVEETV 451
> 7300850
Length=470
Score = 94.4 bits (233), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/65 (66%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
AST+QILGAEKALFRALKT+SNTPKYG+IYH++ +G+A K KG+ISR LA K S+ R+
Sbjct 298 ASTVQILGAEKALFRALKTRSNTPKYGLIYHSSFIGRAGLKNKGRISRFLANKCSIASRI 357
Query 66 DALTE 70
D E
Sbjct 358 DCFLE 362
> CE06114
Length=486
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 41/65 (63%), Positives = 54/65 (83%), Gaps = 0/65 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
AST+QILGAEKALFRALKT+SNTPKYG+++H++ +G+A K KG++SR LA K S+ RV
Sbjct 329 ASTVQILGAEKALFRALKTRSNTPKYGLLFHSSFIGKAGTKNKGRVSRYLANKCSIAARV 388
Query 66 DALTE 70
D +E
Sbjct 389 DCFSE 393
> Hs5453794_1
Length=498
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 41/66 (62%), Positives = 52/66 (78%), Gaps = 0/66 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
AST+QILGAEKALFRALKT+ NTPKYG+I+H+ +G+A K KG+ISR LA K S+ R+
Sbjct 329 ASTVQILGAEKALFRALKTRGNTPKYGLIFHSTFIGRAAAKNKGRISRYLANKCSIASRI 388
Query 66 DALTEA 71
D +E
Sbjct 389 DCFSEV 394
> YLR197w
Length=504
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/66 (63%), Positives = 53/66 (80%), Gaps = 0/66 (0%)
Query 5 SASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVR 64
+AST+QILGAEKALFRALKTK NTPKYG+IYH+ + +A+ K KG+ISR LA K S+ R
Sbjct 329 AASTVQILGAEKALFRALKTKGNTPKYGLIYHSGFISKASAKNKGRISRYLANKCSMASR 388
Query 65 VDALTE 70
+D +E
Sbjct 389 IDNYSE 394
> At1g56110
Length=522
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 40/70 (57%), Positives = 55/70 (78%), Gaps = 0/70 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
+ST+QILGAEKALFRALKT+ NTPKYG+I+H++ +G+A+ K KG+I+R LA K S+ R+
Sbjct 326 SSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGRIARYLANKCSIASRI 385
Query 66 DALTEAAEVA 75
D + A A
Sbjct 386 DCFADGATTA 395
> At3g12860
Length=477
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 39/70 (55%), Positives = 56/70 (80%), Gaps = 0/70 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
+ST+QILGAEKALFRALKT+ NTPKYG+I+H++ +G+A+ K KG+I+R LA K S+ R+
Sbjct 326 SSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGRIARFLANKCSIASRI 385
Query 66 DALTEAAEVA 75
D ++ + A
Sbjct 386 DCFSDNSTTA 395
> At5g27140
Length=445
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 65/166 (39%), Positives = 82/166 (49%), Gaps = 31/166 (18%)
Query 7 STIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRVD 66
STIQILGAEK L++ALKTK TPKYG+IYHA LV QA P+ KGKI+R LAAK +L +R D
Sbjct 282 STIQILGAEKTLYKALKTKQATPKYGLIYHAPLVRQAAPENKGKIARSLAAKSALAIRCD 341
Query 67 ALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAIACRRYVENRLEQLEQQLAGS---- 122
A NG + T+ + R +E RL LE G+
Sbjct 342 AF---------------GNGQ---------DNTMGVESRLKLEARLRNLEGGDLGACEEE 377
Query 123 ---GPKPPSKPAFQRYEPHRETNGVSKKYDVSTDAVDTAAVGSKQK 165
K K A EP E +K + + V+ A SKQ+
Sbjct 378 EEVNDKDTKKEADDEEEPKTEECSKKRKKEAELETVEDPAKKSKQE 423
> ECU04g0820
Length=413
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 36/64 (56%), Positives = 53/64 (82%), Gaps = 0/64 (0%)
Query 7 STIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRVD 66
S+IQ++GAEKALF+ALK+++NTPKYGIIY +L+GQ + KGKI+R LA+K+++ R+D
Sbjct 294 SSIQMMGAEKALFQALKSRTNTPKYGIIYGCSLLGQVPSQHKGKIARSLASKIAIAARID 353
Query 67 ALTE 70
+ E
Sbjct 354 SYGE 357
> ECU09g1600
Length=492
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 46/64 (71%), Gaps = 0/64 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
AST Q+ GAEK+LFR+LK K TPKYG+IY ++ + +A + KG+ISR +A K S+ ++
Sbjct 302 ASTFQLFGAEKSLFRSLKMKKKTPKYGLIYTSSYLSRARDEDKGRISRYIATKCSIAAKI 361
Query 66 DALT 69
D +
Sbjct 362 DCFS 365
> Hs17448109_1
Length=104
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/48 (45%), Positives = 32/48 (66%), Gaps = 0/48 (0%)
Query 24 TKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRVDALTEA 71
TK NTPKYG+++H+ GQA + KG+IS+ LA K S+ ++D E
Sbjct 2 TKGNTPKYGLVFHSTFTGQAAARNKGRISQYLANKCSIASQIDCFAEV 49
> CE24126
Length=504
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 38/67 (56%), Gaps = 0/67 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
+ +Q+LG K T S P +G IY LV + P LK K +++LAAK++L R+
Sbjct 255 SCNVQVLGKTKKNLIGFSTVSTNPHHGFIYFHQLVQKMPPDLKNKAAKILAAKVTLVARI 314
Query 66 DALTEAA 72
DA E++
Sbjct 315 DAQHESS 321
> Hs7661654
Length=499
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 0/68 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
A I +LGA++ + S P G IYH+ +V P L+ K +R++AAK +L RV
Sbjct 246 ACNIMLLGAQRKTLSGFSSTSVLPHTGYIYHSDIVQSLPPDLRRKAARLVAAKCTLAARV 305
Query 66 DALTEAAE 73
D+ E+ E
Sbjct 306 DSFHESTE 313
> 7294306
Length=501
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
A +Q+LGA+K P G +Y++ +V P L+ K +R++AAK L RV
Sbjct 254 ACNVQVLGAQKKTLSGFSQTQMLPHTGYVYYSQIVQDTAPDLRRKAARLVAAKSVLAARV 313
Query 66 DALTEAA 72
DA E+
Sbjct 314 DACHESV 320
> SPBC119.13c
Length=518
Score = 33.5 bits (75), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 31 YGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRVDALTE 70
YG +Y + +V + P ++ + R+ AAK++L R+D++ E
Sbjct 295 YGFLYMSEIVQKTPPDVRKQAIRMTAAKVALAARIDSIHE 334
> At3g60610
Length=442
Score = 32.0 bits (71), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 32/65 (49%), Gaps = 0/65 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
A +Q+LG ++ + ++ + G + + P L+ + R++AAK +L RV
Sbjct 209 ACNVQVLGHKRKNLVGFSSATSQSRVGYLEQTEIFQSTPPGLEARAGRLVAAKSTLAARV 268
Query 66 DALTE 70
DA E
Sbjct 269 DATRE 273
> 7293461
Length=203
Score = 31.6 bits (70), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 148 YDVSTDAVDTAAVGSKQKRHVNGDKGETETQKKKKKVKT 186
Y +S +VD A+VGS Q+ V G G++ +V+T
Sbjct 28 YSISAPSVDHASVGSTQEHTVKGHYGQSSQSDYASQVQT 66
> ECU10g0200
Length=260
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 35/70 (50%), Gaps = 9/70 (12%)
Query 12 LGAEKALFRALKTKSNTPKYGIIYHAA-------LVGQATPKLKGKISRVLAAKLSLCVR 64
+G +L RA T S KYGI + + LV +A+PK +GK+ R L K ++ R
Sbjct 127 MGCLFSLVRA--TPSEVMKYGIKTYGSPLLSQHPLVAKASPKNQGKLLRKLCCKTTIAAR 184
Query 65 VDALTEAAEV 74
+D E+
Sbjct 185 LDFCESNFEI 194
> 7299483
Length=686
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 27/49 (55%), Gaps = 10/49 (20%)
Query 5 SASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISR 53
S +T QIL EKAL KY +I ++ L G+A P++K K S+
Sbjct 181 STTTEQILLTEKAL----------QKYVVITNSPLRGEAQPEIKDKTSK 219
> 7299484
Length=1126
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 27/49 (55%), Gaps = 10/49 (20%)
Query 5 SASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISR 53
S +T QIL EKAL KY +I ++ L G+A P++K K S+
Sbjct 170 STTTEQILLTEKAL----------QKYVVITNSPLRGEAQPEIKDKTSK 208
> At1g60170
Length=511
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 6 ASTIQILGAEKALFRALKTKSNTPKYGIIYHAALVGQATPKLKGKISRVLAAKLSLCVRV 65
A +Q+LG ++ + ++ + G + + P L+ + R++AAK +L RV
Sbjct 273 ACNVQVLGHKRKNLAGFSSATSQSRVGYLEQTEIYQSTPPGLQARAGRLVAAKSTLAARV 332
Query 66 DA 67
DA
Sbjct 333 DA 334
> Hs4557729
Length=1159
Score = 29.3 bits (64), Expect = 4.6, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 43 ATPKLKGKISRVLAAKLSLCVRVDALTE-AAEVAAAAAGGSAANGNSAAAPQGP 95
+ +L+G SR KLS R D TE EV+A G + A +S P GP
Sbjct 873 GSTELEGGFSRQRKRKLSFRRRTDKDTEQPGEVSALGPGRAGAGPSSRGRPGGP 926
> Hs20357498
Length=5171
Score = 28.9 bits (63), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 16/95 (16%)
Query 12 LGAEKALFRALKT--KSNTPKYGII---YHAALVGQA---TPKLKGK-------ISRVLA 56
L A + L ++LK+ K T K I+ + A +G++ T KL+ K I +V
Sbjct 1962 LDAFQVLVKSLKSWIKETTKKVPIVQPSFGAEDLGKSLEDTKKLQEKWSLKTPEIQKVNN 2021
Query 57 AKLSLCVRVDALTEAAE-VAAAAAGGSAANGNSAA 90
+ +SLC + A+T A+ +AA +GG+ NG A
Sbjct 2022 SGISLCNLISAVTTPAKAIAAVKSGGAVLNGEGTA 2056
> 7292835
Length=272
Score = 28.9 bits (63), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 62 CVRVDALTEAAEVAAAAAGGSAANGNSAAAPQGPAEPTVAI 102
C VD L+ A+ + GG +NG SA+A G A VA+
Sbjct 125 CEHVDKLSYASRHKSKLEGGERSNGGSASATTGGANDGVAL 165
> At4g05600
Length=275
Score = 28.5 bits (62), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 96 AEPTVAIACRRYVENRLEQLEQQLAGSGPKPPSKPA 131
E T+AI + NR+E++ + PKPP+KP+
Sbjct 109 METTLAIKEDKRFRNRMEEIIPTYSPEVPKPPTKPS 144
Lambda K H
0.307 0.123 0.331
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3166472848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40