bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1391_orf1
Length=275
Score E
Sequences producing significant alignments: (Bits) Value
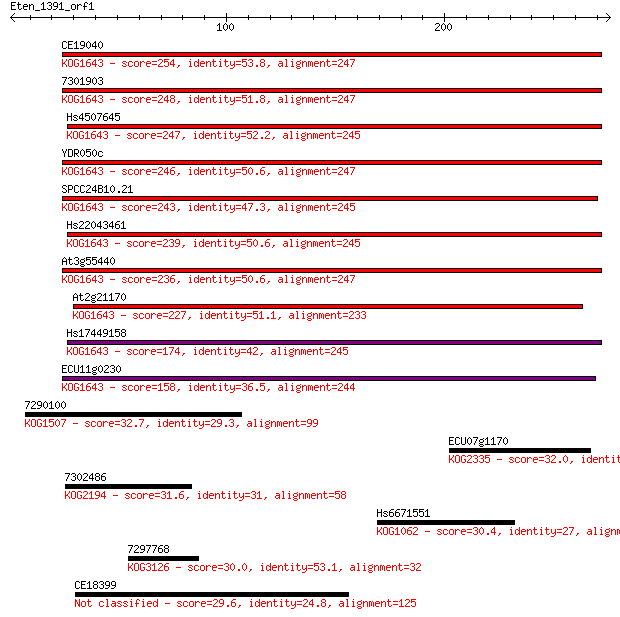
CE19040 254 1e-67
7301903 248 1e-65
Hs4507645 247 2e-65
YDR050c 246 5e-65
SPCC24B10.21 243 4e-64
Hs22043461 239 6e-63
At3g55440 236 5e-62
At2g21170 227 2e-59
Hs17449158 174 2e-43
ECU11g0230 158 1e-38
7290100 32.7 0.74
ECU07g1170 32.0 1.5
7302486 31.6 1.7
Hs6671551 30.4 4.2
7297768 30.0 6.0
CE18399 29.6 7.0
> CE19040
Length=247
Score = 254 bits (649), Expect = 1e-67, Method: Compositional matrix adjust.
Identities = 133/247 (53%), Positives = 166/247 (67%), Gaps = 2/247 (0%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQ 84
M R ++GGNWK NG AS+ + LN A D S +VVV P A ++ AK KLK G
Sbjct 1 MTRKFFVGGNWKMNGDYASVDGIVTFLN-ASADNSSVDVVVAPPAPYLAYAKSKLKAGVL 59
Query 85 IGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEE 144
+ QN K GAFTGEI+ M+KD GL+WV++GHSERR + E+D+++AEK A+ E
Sbjct 60 VAAQNCYKVPKGAFTGEISPAMIKDLGLEWVILGHSERRHVFGESDALIAEKTVHAL-EA 118
Query 145 GIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPE 204
GI V CIGEKLEEREA T +V +QL+A++ K W +VIAYEPVWAIGTGK A+ E
Sbjct 119 GIKVVFCIGEKLEEREAGHTKDVNFRQLQAIVDKGVSWENIVIAYEPVWAIGTGKTASGE 178
Query 205 QAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKA 264
QAQEVH +R F+ EK AVA+ RI+YGGSV+ N L + D+DGFLVGGASLK
Sbjct 179 QAQEVHEWIRAFLKEKVSPAVADATRIIYGGSVTADNAAELGKKPDIDGFLVGGASLKPD 238
Query 265 FVDIIGA 271
FV II A
Sbjct 239 FVKIINA 245
> 7301903
Length=247
Score = 248 bits (633), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 128/247 (51%), Positives = 168/247 (68%), Gaps = 2/247 (0%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQ 84
M R +GGNWK NG SI E++K L+ A DP+ TEVV+ A+++ A+ L
Sbjct 1 MSRKFCVGGNWKMNGDQKSIAEIAKTLSSAALDPN-TEVVIGCPAIYLMYARNLLPCELG 59
Query 85 IGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEE 144
+ QN K GAFTGEI+ M+KD G WV++GHSERR + E+D+++AEK A+ E
Sbjct 60 LAGQNAYKVAKGAFTGEISPAMLKDIGADWVILGHSERRAIFGESDALIAEKAEHALAE- 118
Query 145 GIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPE 204
G+ V CIGE LEEREA KT EV +Q+ A K+ DW+ VV+AYEPVWAIGTG+ ATP+
Sbjct 119 GLKVIACIGETLEEREAGKTNEVVARQMCAYAQKIKDWKNVVVAYEPVWAIGTGQTATPD 178
Query 205 QAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKA 264
QAQEVHA LR+++++ + V+ LRI YGGSV+ N + L + D+DGFLVGGASLK
Sbjct 179 QAQEVHAFLRQWLSDNISKEVSASLRIQYGGSVTAANAKELAKKPDIDGFLVGGASLKPE 238
Query 265 FVDIIGA 271
FVDII A
Sbjct 239 FVDIINA 245
> Hs4507645
Length=249
Score = 247 bits (630), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 128/245 (52%), Positives = 163/245 (66%), Gaps = 2/245 (0%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIG 86
R ++GGNWK NG S+ EL LN A+ P+ TEVV P +I A++KL +
Sbjct 5 RKFFVGGNWKMNGRKQSLGELIGTLNAAKV-PADTEVVCAPPTAYIDFARQKLDPKIAVA 63
Query 87 CQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGI 146
QN K GAFTGEI+ GM+KD G WV++GHSERR + E+D ++ +KVA A+ E G+
Sbjct 64 AQNCYKVTNGAFTGEISPGMIKDCGATWVVLGHSERRHVFGESDELIGQKVAHALAE-GL 122
Query 147 HVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQA 206
V CIGEKL+EREA T +V +Q + + V DW +VV+AYEPVWAIGTGK ATP+QA
Sbjct 123 GVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQA 182
Query 207 QEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAFV 266
QEVH LR ++ +AVA+ RI+YGGSV+ C+ L DVDGFLVGGASLK FV
Sbjct 183 QEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPEFV 242
Query 267 DIIGA 271
DII A
Sbjct 243 DIINA 247
> YDR050c
Length=248
Score = 246 bits (627), Expect = 5e-65, Method: Compositional matrix adjust.
Identities = 125/248 (50%), Positives = 164/248 (66%), Gaps = 3/248 (1%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKN-GF 83
M RT ++GGN+K NG+ SI E+ + LN A P EVV+ P A ++ + +K
Sbjct 1 MARTFFVGGNFKLNGSKQSIKEIVERLNTASI-PENVEVVICPPATYLDYSVSLVKKPQV 59
Query 84 QIGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKE 143
+G QN GAFTGE + +KD G KWV++GHSERR Y+ E D +A+K A+ +
Sbjct 60 TVGAQNAYLKASGAFTGENSVDQIKDVGAKWVILGHSERRSYFHEDDKFIADKTKFALGQ 119
Query 144 EGIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATP 203
G+ V +CIGE LEE++A KT++V +QL AVL +V DW VV+AYEPVWAIGTG ATP
Sbjct 120 -GVGVILCIGETLEEKKAGKTLDVVERQLNAVLEEVKDWTNVVVAYEPVWAIGTGLAATP 178
Query 204 EQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKK 263
E AQ++HA++RKF+A K G+ A ELRI+YGGS + N DVDGFLVGGASLK
Sbjct 179 EDAQDIHASIRKFLASKLGDKAASELRILYGGSANGSNAVTFKDKADVDGFLVGGASLKP 238
Query 264 AFVDIIGA 271
FVDII +
Sbjct 239 EFVDIINS 246
> SPCC24B10.21
Length=249
Score = 243 bits (619), Expect = 4e-64, Method: Compositional matrix adjust.
Identities = 116/245 (47%), Positives = 166/245 (67%), Gaps = 1/245 (0%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQ 84
M R ++GGN+K NG++ S+ + + LN + + E V+FP +++ ++++K
Sbjct 1 MARKFFVGGNFKMNGSLESMKTIIEGLNTTKLNVGDVETVIFPQNMYLITTRQQVKKDIG 60
Query 85 IGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEE 144
+G QN+ GA+TGE +A + D G+ + + GHSERR + E+D VA+K A+ E+
Sbjct 61 VGAQNVFDKKNGAYTGENSAQSLIDAGITYTLTGHSERRTIFKESDEFVADKTKFAL-EQ 119
Query 145 GIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPE 204
G+ V CIGE L EREAN+T+ V +QL A+ KV +W ++VIAYEPVWAIGTGK ATPE
Sbjct 120 GLTVVACIGETLAEREANETINVVVRQLNAIADKVQNWSKIVIAYEPVWAIGTGKTATPE 179
Query 205 QAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKA 264
QAQEVHA +RK+ K G +VAE LR++YGGSV+ NC+ L+ D+DGFLVGGASLK
Sbjct 180 QAQEVHAEIRKWATNKLGASVAEGLRVIYGGSVNGGNCKEFLKFHDIDGFLVGGASLKPE 239
Query 265 FVDII 269
F +I+
Sbjct 240 FHNIV 244
> Hs22043461
Length=249
Score = 239 bits (609), Expect = 6e-63, Method: Compositional matrix adjust.
Identities = 124/245 (50%), Positives = 160/245 (65%), Gaps = 2/245 (0%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIG 86
R ++GGNWK NG S+ EL + LN A+ P+ TEVV P +I A++KL +
Sbjct 5 RKFFVGGNWKMNGRKQSLRELVRTLNAAKV-PADTEVVCTPPTAYIDFARQKLDPKIAVA 63
Query 87 CQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGI 146
QN K GAFTGEI+ GM+KD WV++GHSERR + E+D ++ +KVA A+ E G+
Sbjct 64 AQNCYKVTNGAFTGEISPGMIKDCRATWVVLGHSERRHVFGESDELIGQKVAHALAE-GL 122
Query 147 HVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQA 206
V C GEKL+EREA T +V +Q + + V DW +VV+AYEPVWAIGTGK ATP+Q
Sbjct 123 GVIACTGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQD 182
Query 207 QEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAFV 266
QEVH LR ++ +AVA+ RI+YGGSV+ C+ L VDGFLVGGASLK FV
Sbjct 183 QEVHDKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPGVDGFLVGGASLKPEFV 242
Query 267 DIIGA 271
DII A
Sbjct 243 DIINA 247
> At3g55440
Length=254
Score = 236 bits (601), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 125/249 (50%), Positives = 164/249 (65%), Gaps = 4/249 (1%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQ--TEVVVFPTALHIGLAKEKLKNG 82
M R ++GGNWK NGT + ++ LN A+ PSQ EVVV P + + L K L++
Sbjct 1 MARKFFVGGNWKCNGTAEEVKKIVNTLNEAQV-PSQDVVEVVVSPPYVFLPLVKSTLRSD 59
Query 83 FQIGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMK 142
F + QN GAFTGE++A M+ + + WV++GHSERR +E+ V +KVA A+
Sbjct 60 FFVAAQNCWVKKGGAFTGEVSAEMLVNLDIPWVILGHSERRAILNESSEFVGDKVAYALA 119
Query 143 EEGIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVAT 202
+ G+ V C+GE LEEREA TM+V Q +A+ +V++W VVIAYEPVWAIGTGKVA+
Sbjct 120 Q-GLKVIACVGETLEEREAGSTMDVVAAQTKAIADRVTNWSNVVIAYEPVWAIGTGKVAS 178
Query 203 PEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLK 262
P QAQEVH LRK++A+ VA RI+YGGSV+ NC+ L DVDGFLVGGASLK
Sbjct 179 PAQAQEVHDELRKWLAKNVSADVAATTRIIYGGSVNGGNCKELGGQADVDGFLVGGASLK 238
Query 263 KAFVDIIGA 271
F+DII A
Sbjct 239 PEFIDIIKA 247
> At2g21170
Length=315
Score = 227 bits (578), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 119/233 (51%), Positives = 152/233 (65%), Gaps = 2/233 (0%)
Query 30 WIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIGCQN 89
++GGNWK NGT SI +L +LN A + + +VVV P ++I K L + I QN
Sbjct 67 FVGGNWKCNGTKDSIAKLISDLNSATLE-ADVDVVVSPPFVYIDQVKSSLTDRIDISGQN 125
Query 90 ISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGIHVT 149
GAFTGEI+ +KD G KWV++GHSERR E D + +K A A+ EG+ V
Sbjct 126 SWVGKGGAFTGEISVEQLKDLGCKWVILGHSERRHVIGEKDEFIGKKAAYAL-SEGLGVI 184
Query 150 ICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQAQEV 209
CIGEKLEEREA KT +VC QL+A V W +V+AYEPVWAIGTGKVA+P+QAQEV
Sbjct 185 ACIGEKLEEREAGKTFDVCFAQLKAFADAVPSWDNIVVAYEPVWAIGTGKVASPQQAQEV 244
Query 210 HAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLK 262
H A+R ++ + E VA + RI+YGGSV+ N L + D+DGFLVGGASLK
Sbjct 245 HVAVRGWLKKNVSEEVASKTRIIYGGSVNGGNSAELAKEEDIDGFLVGGASLK 297
> Hs17449158
Length=227
Score = 174 bits (441), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 103/245 (42%), Positives = 136/245 (55%), Gaps = 24/245 (9%)
Query 27 RTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALHIGLAKEKLKNGFQIG 86
R ++G NWK NG + EL N A P+ T+V+ + LA++KL +
Sbjct 5 RKFFVGRNWKMNGRKKCLGELIGTQNAATV-PADTKVICALATAYNELARQKLAPKIAVA 63
Query 87 CQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKEEGI 146
QN K GAFTGEI+ GMVKD G+ KVA A+ E +
Sbjct 64 PQNCYKVTNGAFTGEISPGMVKDLGV----------------------TKVAHALAER-L 100
Query 147 HVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATPEQA 206
V CIGEKL+EREA T +V +Q + + V DW +V++AY+PVWA GTGK ATP+QA
Sbjct 101 GVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVILAYDPVWATGTGKTATPQQA 160
Query 207 QEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKKAFV 266
QEVH LR ++ +AVA+ I+YGGSV++ C+ L DV FL+ G SLK FV
Sbjct 161 QEVHKKLRGWLKSNISDAVAQSTGIIYGGSVTKATCKELASQPDVAAFLMSGVSLKPEFV 220
Query 267 DIIGA 271
DII A
Sbjct 221 DIINA 225
> ECU11g0230
Length=242
Score = 158 bits (399), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 89/245 (36%), Positives = 147/245 (60%), Gaps = 12/245 (4%)
Query 25 MPRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVV-FPTALHIGLAKEKLKNGF 83
M + GNWK + ++ + E+ K F + T + V FP +I +AK+
Sbjct 1 MKGNKLLAGNWKMSSSLQTF-EIIKSFRANLFAGNDTFIAVPFP---YIHVAKKMFPEYI 56
Query 84 QIGCQNISKTGPGAFTGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAEKVAMAMKE 143
++G Q+ S+ G +TGE++A MV + G ++V++GHSERR+++ ET + V +K+ A +
Sbjct 57 KVGAQDCSQFKSGPYTGEVSASMVSEMGAEYVIIGHSERRRWFGETSATVVKKIRNA-RS 115
Query 144 EGIHVTICIGEKLEEREANKTMEVCRQQLEAVLPKVSDWRRVVIAYEPVWAIGTGKVATP 203
EG+ + +CIGE + RE + ++V + QL ++ ++ + IAYEPVWAIGTGKV T
Sbjct 116 EGLKIVVCIGEDEKCREDGRHLDVIKDQLLSLGEEIGG-EGIDIAYEPVWAIGTGKVPTN 174
Query 204 EQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASLKK 263
+Q +EV + ++++ G + R++YGGSVS NC+ +L ++DGFL+GG SLK
Sbjct 175 DQIREVVSGIKRW-----GTGMGFVGRVIYGGSVSMLNCQTILDVEEIDGFLIGGTSLKM 229
Query 264 AFVDI 268
F I
Sbjct 230 DFCMI 234
> 7290100
Length=362
Score = 32.7 bits (73), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 48/112 (42%), Gaps = 13/112 (11%)
Query 8 LCSSAEPHIPFALAAAKMPRTPWIGGNWKSNGTVASITELSKELNHA----EFDPSQTEV 63
L SA+P PF ++ R +W+ + T S+ N A + P ++
Sbjct 231 LQHSADPEYPFLFEGPEIVRCEGCHIHWRDGSNLTLQTVESRRRNRAHRVTKVMPRESFF 290
Query 64 VVF--PTALHIGLAKEKLK----NGFQIGCQNISKTGPGA---FTGEITAGM 106
F P AL + LA EK K N F++G ++ P A +TG++ +
Sbjct 291 RFFAPPQALDLSLADEKTKLILGNDFEVGFLLRTQIVPKAVLFYTGDLVDSL 342
> ECU07g1170
Length=357
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 37/65 (56%), Gaps = 5/65 (7%)
Query 202 TPEQAQEVHAALRKFVAEKAGEAVAEELRIVYGGSVSEQNCEALLRCTDVDGFLVGGASL 261
+P+Q ++ A + + A +++ ELRI+ GS+ E N + + ++DG ++G ++
Sbjct 185 SPQQNRK--APPLDYASVYAIKSLHPELRIILNGSIGEGNLDKI---HNLDGVMIGRGAI 239
Query 262 KKAFV 266
+ FV
Sbjct 240 RNVFV 244
> 7302486
Length=765
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Query 26 PRTPWIGGNWKSNGTVASITELSKELNHAEFDPSQTEVVVFPTALH-IGLAKEKLKNGF 83
P +PW+ +K N T +++E+ PS T+ +F + IGL K NGF
Sbjct 152 PNSPWLVEKYKENAPHYLATTMAEEIFQTGILPSDTDFAIFVKYGNLIGLDMAKFINGF 210
> Hs6671551
Length=1137
Score = 30.4 bits (67), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 31/66 (46%), Gaps = 3/66 (4%)
Query 169 RQQLEAVLPKVSD---WRRVVIAYEPVWAIGTGKVATPEQAQEVHAALRKFVAEKAGEAV 225
R+ + AVLP + D + ++ + V A+ + P Q Q +H RK + ++ +
Sbjct 163 REMIPAVLPLIEDKLQHSKEIVRRKAVLALYKFHLIAPNQVQHIHIKFRKALCDRDVGVM 222
Query 226 AEELRI 231
A L I
Sbjct 223 AASLHI 228
> 7297768
Length=358
Score = 30.0 bits (66), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Query 55 EFDPSQTEVVVFPTALHIG-LAKEKLKNGFQIG 86
E PS T PT H+G LAK+ L NGF+IG
Sbjct 68 EIIPSPTSEGEMPTYFHVGALAKDCLINGFKIG 100
> CE18399
Length=307
Score = 29.6 bits (65), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 31/140 (22%), Positives = 52/140 (37%), Gaps = 26/140 (18%)
Query 31 IGGNWKSNGTVASITELSKELNHAEFDPSQTE--VVVFPTALHIGLAKEKLKNGFQIGCQ 88
I G +N + S+ELNH F+ S ++ + F ++ L LK+G IG
Sbjct 50 IYGGLSTNDNFTRYLQFSRELNHVTFEGSTSDDRLTFFINVYNMMLIHITLKHGPPIGIW 109
Query 89 NISKTGPGAF-------------TGEITAGMVKDFGLKWVMVGHSERRQYYSETDSVVAE 135
K G + I G K G+ W G + R
Sbjct 110 QRRKLVNGTYYLIGGHRYALHSIINGILRGNKKGPGMLWKAFGKQDARL----------- 158
Query 136 KVAMAMKEEGIHVTICIGEK 155
+++A+ + I+ ++C G K
Sbjct 159 PISLAVCDPLIYFSLCSGSK 178
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5950231728
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40