bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1491_orf2
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
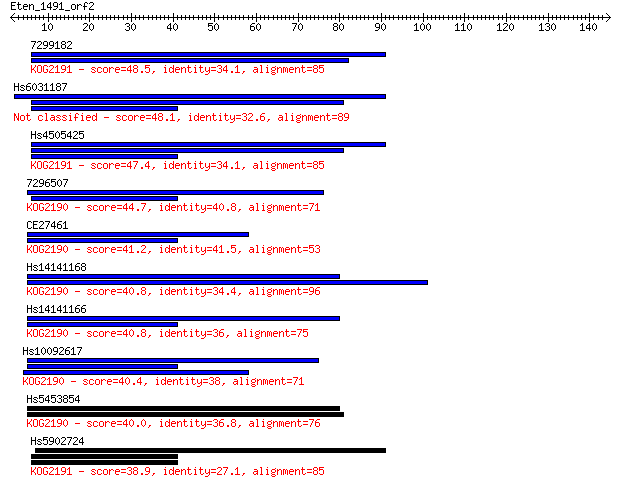
7299182 48.5 4e-06
Hs6031187 48.1 6e-06
Hs4505425 47.4 1e-05
7296507 44.7 6e-05
CE27461 41.2 7e-04
Hs14141168 40.8 9e-04
Hs14141166 40.8 0.001
Hs10092617 40.4 0.001
Hs5453854 40.0 0.001
Hs5902724 38.9 0.003
> 7299182
Length=493
Score = 48.5 bits (114), Expect = 4e-06, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 46/87 (52%), Gaps = 7/87 (8%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS--PLFVTAEEACAERIVSVESRKRQ 63
V+I VP + G +IGK G +I+ ++ +G+ + IS P V+ +E C I ++
Sbjct 94 VKILVPNSTAGMIIGKGGAFIKQIKEESGSYVQISQKPTDVSLQERCITII-----GDKE 148
Query 64 NLKAAAFTLIKKINEHPDKGCCRHVCY 90
N K A ++ KI E P G C +V Y
Sbjct 149 NNKNACKMILSKIVEDPQSGTCLNVSY 175
Score = 32.0 bits (71), Expect = 0.44, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 35/76 (46%), Gaps = 2/76 (2%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQNL 65
++I VP G++IGK G I SL+ TGA + +S ER+ + +
Sbjct 1 MKILVPAVASGAIIGKGGETIASLQKDTGARVKMSKSH-DFYPGTTERVCLITGSTEAIM 59
Query 66 KAAAFTLIKKINEHPD 81
F ++ KI E PD
Sbjct 60 VVMEF-IMDKIREKPD 74
> Hs6031187
Length=486
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 49/91 (53%), Gaps = 7/91 (7%)
Query 2 QLSTVRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS--PLFVTAEEACAERIVSVES 59
++ V+I VP + G +IGK G ++++ +GA + +S P + + ER+V+V
Sbjct 149 RIKQVKIIVPNSTAGLIIGKGGATVKAVMEQSGAWVQLSQKPDGINLQ----ERVVTVSG 204
Query 60 RKRQNLKAAAFTLIKKINEHPDKGCCRHVCY 90
QN KA +I+KI E P G C ++ Y
Sbjct 205 EPEQNRKAVEL-IIQKIQEDPQSGSCLNISY 234
Score = 34.3 bits (77), Expect = 0.081, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 3/77 (3%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAE--EACAERIVSVESRKRQ 63
+++ +P GS+IGK G I L+ TGATI +S L + + ER+ ++ +
Sbjct 52 LKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKLSKSKDFYPGTTERVCLIQG-TVE 110
Query 64 NLKAAAFTLIKKINEHP 80
L A + +KI E P
Sbjct 111 ALNAVHGFIAEKIREMP 127
Score = 29.6 bits (65), Expect = 2.0, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
V I VP +VG+++GK G + + TGA I IS
Sbjct 403 VEIAVPENLVGAILGKGGKTLVEYQELTGARIQIS 437
> Hs4505425
Length=510
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 47/87 (54%), Gaps = 7/87 (8%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS--PLFVTAEEACAERIVSVESRKRQ 63
V+I VP + G +IGK G ++++ +GA + +S P + + ER+V+V Q
Sbjct 177 VKIIVPNSTAGLIIGKGGATVKAVMEQSGAWVQLSQKPDGINLQ----ERVVTVSGEPEQ 232
Query 64 NLKAAAFTLIKKINEHPDKGCCRHVCY 90
N KA +I+KI E P G C ++ Y
Sbjct 233 NRKAVEL-IIQKIQEDPQSGSCLNISY 258
Score = 34.3 bits (77), Expect = 0.084, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 3/77 (3%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAE--EACAERIVSVESRKRQ 63
+++ +P GS+IGK G I L+ TGATI +S L + + ER+ ++ +
Sbjct 52 LKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKLSKSKDFYPGTTERVCLIQG-TVE 110
Query 64 NLKAAAFTLIKKINEHP 80
L A + +KI E P
Sbjct 111 ALNAVHGFIAEKIREMP 127
Score = 29.6 bits (65), Expect = 2.0, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
V I VP +VG+++GK G + + TGA I IS
Sbjct 427 VEIAVPENLVGAILGKGGKTLVEYQELTGARIQIS 461
> 7296507
Length=386
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQN 64
T+R+ + VGS+IGK G + R +GA INIS + +C ERIV+V N
Sbjct 25 TIRLIMQGKEVGSIIGKKGEIVNRFREESGAKINIS------DGSCPERIVTVSGTT--N 76
Query 65 LKAAAFTLIKK 75
+AFTLI K
Sbjct 77 AIFSAFTLITK 87
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
+R+ VP + GSLIGK+G I+ +R TG +I ++
Sbjct 109 IRLIVPASQCGSLIGKSGSKIKEIRQTTGCSIQVA 143
> CE27461
Length=451
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 33/53 (62%), Gaps = 6/53 (11%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSV 57
T+R+ + VGS+IGK G I+ +R +GA INIS + +C ERIV++
Sbjct 73 TIRLLMQGKEVGSIIGKKGDQIKKIREESGAKINIS------DGSCPERIVTI 119
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
T+R+ VP GSLIGK G I+ +R ATGA+I ++
Sbjct 154 TMRVIVPATQCGSLIGKGGSKIKDIREATGASIQVA 189
> Hs14141168
Length=366
Score = 40.8 bits (94), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQN 64
T+R+ + VGS+IGK G ++ +R +GA INIS E C ERI+++
Sbjct 15 TIRLLMHGKEVGSIIGKKGESVKKMREESGARINIS------EGNCPERIITLAGPTNAI 68
Query 65 LKAAAFTLIKKINEH 79
KA A +I K+ E
Sbjct 69 FKAFAM-IIDKLEED 82
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 46/96 (47%), Gaps = 3/96 (3%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQN 64
T+R+ VP + GSLIGK G I+ +R +TGA + ++ + A I + +
Sbjct 99 TLRLVVPASQCGSLIGKGGCKIKEIRESTGAQVQVAGDMLPNSTERAITIAGIPQSIIEC 158
Query 65 LKAAAFTLIKKINEHPDKGCCRHVCYFRKHSFESPL 100
+K +++ +++ P KG +R SP+
Sbjct 159 VKQICVVMLETLSQSPPKGV---TIPYRPKPSSSPV 191
> Hs14141166
Length=362
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQN 64
T+R+ + VGS+IGK G ++ +R +GA INIS E C ERI+++
Sbjct 15 TIRLLMHGKEVGSIIGKKGESVKKMREESGARINIS------EGNCPERIITLAGPTNAI 68
Query 65 LKAAAFTLIKKINEH 79
KA A +I K+ E
Sbjct 69 FKAFAM-IIDKLEED 82
Score = 32.7 bits (73), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
T+R+ VP + GSLIGK G I+ +R +TGA + ++
Sbjct 99 TLRLVVPASQCGSLIGKGGCKIKEIRESTGAQVQVA 134
> Hs10092617
Length=339
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 36/70 (51%), Gaps = 6/70 (8%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQN 64
T+R+ + VGS+IGK G ++ +R +GA INIS E C ERIV++
Sbjct 15 TIRLLMHGKEVGSIIGKKGETVKKMREESGARINIS------EGNCPERIVTITGPTDAI 68
Query 65 LKAAAFTLIK 74
KA A K
Sbjct 69 FKAFAMIAYK 78
Score = 34.7 bits (78), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
T+R+ VP + GSLIGK G I+ +R +TGA + ++
Sbjct 99 TLRLVVPASQCGSLIGKGGSKIKEIRESTGAQVQVA 134
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 30/54 (55%), Gaps = 4/54 (7%)
Query 4 STVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSV 57
ST + +P ++G +IG+ G I +R +GA I I+ A E +ER +++
Sbjct 262 STHELTIPNDLIGCIIGRQGTKINEIRQMSGAQIKIA----NATEGSSERQITI 311
> Hs5453854
Length=356
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQN 64
T+R+ + VGS+IGK G ++ +R +GA INIS E C ERI+++
Sbjct 15 TIRLLMHGKEVGSIIGKKGESVKRIREESGARINIS------EGNCPERIITLTGPTNAI 68
Query 65 LKAAAFTLIKKINEH 79
KA A +I K+ E
Sbjct 69 FKAFAM-IIDKLEED 82
Score = 33.5 bits (75), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 38/76 (50%), Gaps = 0/76 (0%)
Query 5 TVRICVPRAVVGSLIGKNGGYIQSLRVATGATINISPLFVTAEEACAERIVSVESRKRQN 64
T+R+ VP GSLIGK G I+ +R +TGA + ++ + A I V +
Sbjct 99 TLRLVVPATQCGSLIGKGGCKIKEIRESTGAQVQVAGDMLPNSTERAITIAGVPQSVTEC 158
Query 65 LKAAAFTLIKKINEHP 80
+K +++ +++ P
Sbjct 159 VKQICLVMLETLSQSP 174
> Hs5902724
Length=492
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 45/86 (52%), Gaps = 7/86 (8%)
Query 7 RICVPRAVVGSLIGKNGGYIQSLRVATGATINIS--PLFVTAEEACAERIVSVESRKRQN 64
++ VP + G +IGK G ++++ +GA + +S P + + ER+V+V Q
Sbjct 134 KLIVPNSTAGLIIGKGGATVKAVMEQSGAWVQLSQKPEGINLQ----ERVVTVSGEPEQV 189
Query 65 LKAAAFTLIKKINEHPDKGCCRHVCY 90
KA + +++K+ E P C ++ Y
Sbjct 190 HKAVS-AIVQKVQEDPQSSSCLNISY 214
Score = 32.0 bits (71), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
+++ +P GS+IGK G I L+ TGATI +S
Sbjct 35 LKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLS 69
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 6 VRICVPRAVVGSLIGKNGGYIQSLRVATGATINIS 40
V I VP +VG+++GK G + + TGA I IS
Sbjct 409 VEIAVPENLVGAILGKGGKTLVEYQELTGARIQIS 443
Lambda K H
0.321 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1675978996
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40