bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1504_orf2
Length=63
Score E
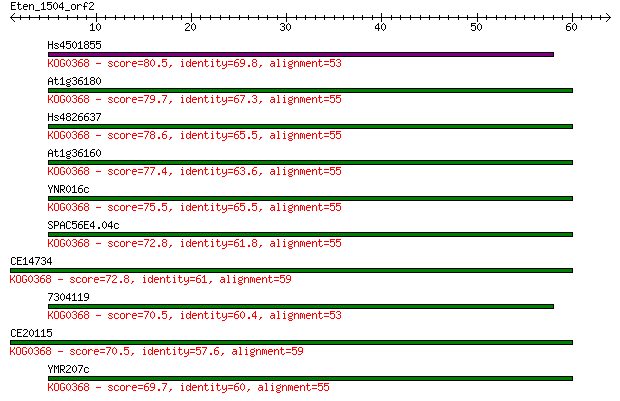
Sequences producing significant alignments: (Bits) Value
Hs4501855 80.5 7e-16
At1g36180 79.7 1e-15
Hs4826637 78.6 2e-15
At1g36160 77.4 6e-15
YNR016c 75.5 2e-14
SPAC56E4.04c 72.8 2e-13
CE14734 72.8 2e-13
7304119 70.5 7e-13
CE20115 70.5 7e-13
YMR207c 69.7 1e-12
> Hs4501855
Length=2483
Score = 80.5 bits (197), Expect = 7e-16, Method: Composition-based stats.
Identities = 37/53 (69%), Positives = 46/53 (86%), Gaps = 0/53 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTS 57
LGVENLRGSG IAGE+SLAY+E+ T+SLV+ R++GIGAY+VRL QR IQ + S
Sbjct 1928 LGVENLRGSGMIAGESSLAYEEIVTISLVTCRAIGIGAYLVRLGQRVIQVENS 1980
> At1g36180
Length=2359
Score = 79.7 bits (195), Expect = 1e-15, Method: Composition-based stats.
Identities = 37/55 (67%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LGVENL GSG IAG S AY+E FTL+ VSGRSVGIGAY+ RL RCIQ+ P+
Sbjct 1814 LGVENLTGSGAIAGAYSRAYNETFTLTFVSGRSVGIGAYLARLGMRCIQRLDQPI 1868
> Hs4826637
Length=2346
Score = 78.6 bits (192), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/55 (65%), Positives = 46/55 (83%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+G ENLRGSG IAGE+SLAY+E+ T+SLV+ R++GIGAY+VRL QR IQ + S L
Sbjct 1792 IGPENLRGSGMIAGESSLAYNEIITISLVTSRAIGIGAYLVRLGQRTIQVENSHL 1846
> At1g36160
Length=2247
Score = 77.4 bits (189), Expect = 6e-15, Method: Composition-based stats.
Identities = 35/55 (63%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+GVENL GSG IAG S AY+E FTL+ VSGR+VGIGAY+ RL RCIQ+ P+
Sbjct 1702 IGVENLTGSGAIAGAYSKAYNETFTLTFVSGRTVGIGAYLARLGMRCIQRLDQPI 1756
> YNR016c
Length=2233
Score = 75.5 bits (184), Expect = 2e-14, Method: Composition-based stats.
Identities = 36/55 (65%), Positives = 44/55 (80%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LGVE LRGSG IAG TS AY ++FT++LV+ RSVGIGAY+VRL QR IQ + P+
Sbjct 1700 LGVECLRGSGLIAGATSRAYHDIFTITLVTCRSVGIGAYLVRLGQRAIQVEGQPI 1754
> SPAC56E4.04c
Length=2280
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 34/55 (61%), Positives = 44/55 (80%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LGVE LRGSG IAG TS AY+++FT +LV+ R+VGIGAY+VRL QR +Q + P+
Sbjct 1741 LGVECLRGSGLIAGVTSRAYNDIFTCTLVTCRAVGIGAYLVRLGQRAVQIEGQPI 1795
> CE14734
Length=2054
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 36/59 (61%), Positives = 43/59 (72%), Gaps = 0/59 (0%)
Query 1 EGQHLGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+ + +GVENL+GSG IAGET+ AY EV T V+GRSVGIGAY RLA R +Q K S L
Sbjct 1543 KNEKIGVENLQGSGLIAGETARAYAEVPTYCYVTGRSVGIGAYTARLAHRIVQHKQSHL 1601
> 7304119
Length=2348
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 32/53 (60%), Positives = 43/53 (81%), Gaps = 0/53 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTS 57
LGVENLR +G IAGETS AY+E+ T+++V+ R++GIG+Y+VRL QR IQ S
Sbjct 1789 LGVENLRYAGLIAGETSQAYEEIVTIAMVTCRTIGIGSYVVRLGQRVIQIDNS 1841
> CE20115
Length=1657
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 34/59 (57%), Positives = 43/59 (72%), Gaps = 0/59 (0%)
Query 1 EGQHLGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+ +++GVENL GSG I GETS AY E+ T V+GRSVGIGAY RLA+R IQ + + L
Sbjct 1223 KNEYIGVENLMGSGAIGGETSRAYREIPTYCYVTGRSVGIGAYTARLARRIIQHEKAHL 1281
> YMR207c
Length=2123
Score = 69.7 bits (169), Expect = 1e-12, Method: Composition-based stats.
Identities = 33/55 (60%), Positives = 43/55 (78%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LGVE L+GSG IAG TS AY ++FT++ V+ RSVGIG+Y+VRL QR IQ + P+
Sbjct 1595 LGVECLQGSGLIAGATSKAYRDIFTITAVTCRSVGIGSYLVRLGQRTIQVEDKPI 1649
Lambda K H
0.321 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1172754882
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40