bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1541_orf1
Length=155
Score E
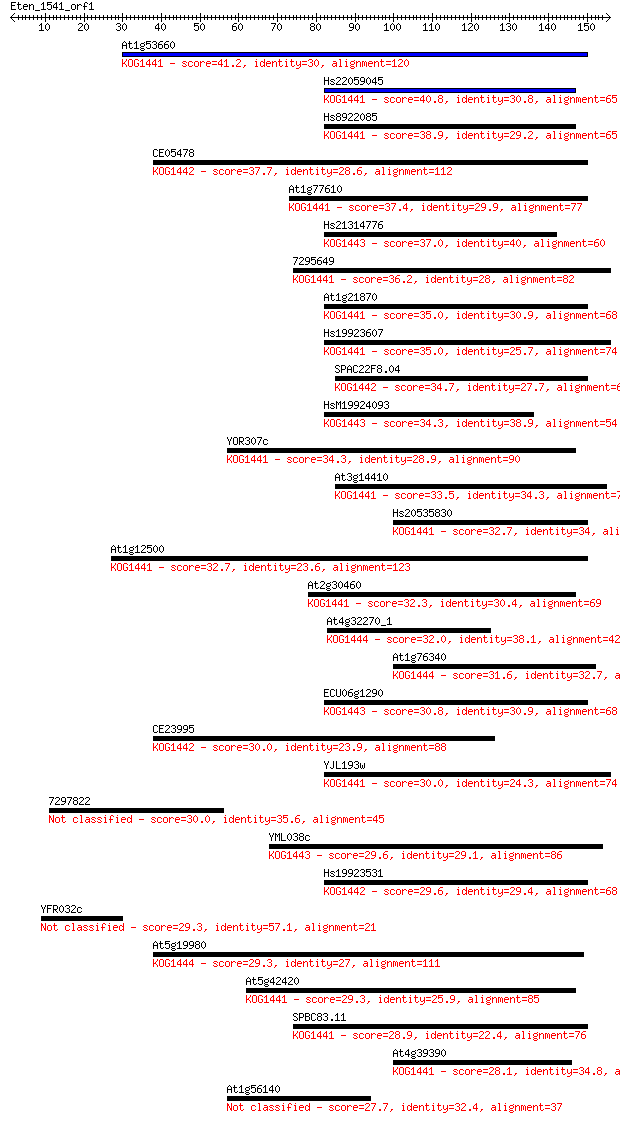
Sequences producing significant alignments: (Bits) Value
At1g53660 41.2 9e-04
Hs22059045 40.8 0.001
Hs8922085 38.9 0.004
CE05478 37.7 0.009
At1g77610 37.4 0.012
Hs21314776 37.0 0.016
7295649 36.2 0.027
At1g21870 35.0 0.056
Hs19923607 35.0 0.057
SPAC22F8.04 34.7 0.080
HsM19924093 34.3 0.088
YOR307c 34.3 0.095
At3g14410 33.5 0.16
Hs20535830 32.7 0.27
At1g12500 32.7 0.29
At2g30460 32.3 0.41
At4g32270_1 32.0 0.43
At1g76340 31.6 0.62
ECU06g1290 30.8 0.96
CE23995 30.0 1.7
YJL193w 30.0 1.8
7297822 30.0 2.1
YML038c 29.6 2.4
Hs19923531 29.6 2.4
YFR032c 29.3 3.0
At5g19980 29.3 3.0
At5g42420 29.3 3.1
SPBC83.11 28.9 4.0
At4g39390 28.1 6.2
At1g56140 27.7 8.4
> At1g53660
Length=316
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 59/124 (47%), Gaps = 15/124 (12%)
Query 30 RAEPLCMFYNM--LNALCLFPIVVLVSEEPKVLWKLAAEGSMMSGTVLAQVVGLG-VVTF 86
+ PL + Y M +A+CLF + W + M + V+ L + TF
Sbjct 179 KLNPLSLMYYMSPCSAICLF-----------IPWIFLEKSKMDTWNFHVLVLSLNSLCTF 227
Query 87 VLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGE-RLTLVETCGFLLTLGGSLWY 145
L S+F +S TS L+ +AG VK L V V+ LL+ E +LT++ G+ + + G Y
Sbjct 228 ALNLSVFLVISRTSALTIRIAGVVKDWLVVLVSALLFAETKLTIINLFGYAVAIVGVATY 287
Query 146 SVSR 149
+ +
Sbjct 288 NNHK 291
> Hs22059045
Length=129
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
GV+ F++ SI+W + TSP+++++ G+ K + + +L+ + L++ + G L TL G
Sbjct 47 GVIAFMVNLSIYWIIGNTSPVTYNMFGHFKFCITLFGGYVLFKDPLSINQALGILCTLFG 106
Query 142 SLWYS 146
L Y+
Sbjct 107 ILAYT 111
> Hs8922085
Length=354
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
GV+ F++ SI+W + TSP+++++ G+ K + + +L+ + L++ + L TL G
Sbjct 272 GVIAFMVNLSIYWIIGNTSPVTYNMFGHFKFCIALFGGYVLFKDPLSINQALDILCTLFG 331
Query 142 SLWYS 146
L Y+
Sbjct 332 ILAYT 336
> CE05478
Length=416
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 55/114 (48%), Gaps = 6/114 (5%)
Query 38 YNMLNALCLF-PIVVLVSEEPKVLWKLAAEGSMMSGTVLAQVVGLGVVTFVLGTSIFWTV 96
YN LNAL LF P+++ E V + + T + GV F++G W +
Sbjct 272 YNNLNALVLFLPLMLFNGEFGAVFY----FDKLFDTTFWILMTLGGVFGFMMGYVTGWQI 327
Query 97 SLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLV-ETCGFLLTLGGSLWYSVSR 149
TSPL+ +++G K Q +A++ + E TL+ T F++ G ++ V +
Sbjct 328 QATSPLTHNISGTAKAAAQTVMAVVWYSELKTLLWWTSNFVVLFGSGMYTYVQK 381
> At1g77610
Length=336
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 73 TVLAQVVGLGVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVET 132
+ L ++ GV+ F L SIF+ + T+ ++F+VAG +K + V V+ L++ ++ +
Sbjct 224 SALIIILSSGVLAFCLNFSIFYVIHSTTAVTFNVAGNLKVAVAVMVSWLIFRNPISYMNA 283
Query 133 CGFLLTLGGSLWYSVSR 149
G +TL G +Y R
Sbjct 284 VGCGITLVGCTFYGYVR 300
> Hs21314776
Length=365
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
G++ F LG S F VS TS L+ S+AG K + + +A L G++++L+ GF L L G
Sbjct 250 GILAFGLGFSEFLLVSRTSSLTLSIAGIFKEVCTLLLAAHLLGDQISLLNWLGFALCLSG 309
> 7295649
Length=373
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 43/82 (52%), Gaps = 0/82 (0%)
Query 74 VLAQVVGLGVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETC 133
V+A + GV+ ++ F +SL +PL+++VA K + + V++L+ G +T V
Sbjct 228 VIALLFADGVLNWLQNIIAFSVLSLVTPLTYAVASASKRIFVIAVSLLILGNPVTWVNCV 287
Query 134 GFLLTLGGSLWYSVSRCQANAR 155
G L + G L Y+ ++ R
Sbjct 288 GMTLAIVGVLCYNRAKQLTRGR 309
> At1g21870
Length=341
Score = 35.0 bits (79), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 39/68 (57%), Gaps = 0/68 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
GV+ F L SIF+ + T+ ++F+VAG +K + V V+ +++ ++ + G +TL G
Sbjct 239 GVLAFCLNFSIFYVIQSTTAVTFNVAGNLKVAVAVFVSWMIFRNPISPMNAVGCGITLVG 298
Query 142 SLWYSVSR 149
+Y R
Sbjct 299 CTFYGYVR 306
> Hs19923607
Length=266
Score = 35.0 bits (79), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
G F F ++L SPLS+SVA K ++ + V++++ +T G + + G
Sbjct 125 GFCNFAQNVIAFSILNLVSPLSYSVANATKRIMVITVSLIMLRNPVTSTNVLGMMTAILG 184
Query 142 SLWYSVSRCQANAR 155
Y+ ++ AN +
Sbjct 185 VFLYNKTKYDANQQ 198
> SPAC22F8.04
Length=383
Score = 34.7 bits (78), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 37/65 (56%), Gaps = 0/65 (0%)
Query 85 TFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGGSLW 144
F L + F + +TSP+++ ++ +++LQ +A+ GE L G +L L G+L
Sbjct 310 NFYLNIATFTQIKVTSPVTYMISVSARSILQTLLAVAFLGETLYGNRIYGVILILVGTLL 369
Query 145 YSVSR 149
Y++++
Sbjct 370 YTLAK 374
> HsM19924093
Length=329
Score = 34.3 bits (77), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGF 135
G++ F LG S F VS TS L+ S+AG K + + +A L G++++L+ GF
Sbjct 250 GILAFGLGFSEFLLVSRTSSLTLSIAGIFKEVCTLLLAAHLLGDQISLLNWLGF 303
> YOR307c
Length=453
Score = 34.3 bits (77), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 44/93 (47%), Gaps = 4/93 (4%)
Query 57 PKVLWKLAAEGSMMSGTVL---AQVVGLGVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTL 113
P + +L GS+++ L A V G+ F F + L S +++SVA +K +
Sbjct 350 PFLTGELMHGGSVINDLTLETVALVAIHGIAHFFQAMLAFQLIGLLSSINYSVANIMKRI 409
Query 114 LQVGVAMLLWGERLTLVETCGFLLTLGGSLWYS 146
+ + VA L W +L + G +LT+ G Y
Sbjct 410 VVISVA-LFWETKLNFFQVFGVILTIAGLYGYD 441
> At3g14410
Length=285
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 1/71 (1%)
Query 85 TFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGE-RLTLVETCGFLLTLGGSL 143
TF L S+F +S TS L+ VAG VK + V V+ LL+ + +LT++ G+ + + G
Sbjct 188 TFALNLSVFLVISHTSALTIRVAGVVKDWVVVLVSALLFADTKLTIINLFGYAIAIAGVA 247
Query 144 WYSVSRCQANA 154
Y+ + + A
Sbjct 248 AYNNHKLKKEA 258
> Hs20535830
Length=417
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 100 SPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGGSLWYSVSR 149
SP++FSVA VK L + ++++++G ++T + G L G L Y+ +R
Sbjct 322 SPVTFSVASTVKHALSIWLSVIVFGNKITSLSAVGTALVTVGVLLYNKAR 371
> At1g12500
Length=361
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 29/123 (23%), Positives = 59/123 (47%), Gaps = 3/123 (2%)
Query 27 ERPRAEPLCMFYNMLNALCLFPIVVLVSEEPKVLWKLAAEGSMMSGTVLAQVVGLGVVTF 86
E+ + L ++ + A L P + + E VL ++ E + ++ + G V +
Sbjct 233 EKLHSMNLLLYMAPMAACILLPFTLYI--EGNVL-RVLIEKARTDPLIIFLLAGNATVAY 289
Query 87 VLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGGSLWYS 146
++ + F TS L+ V G K + GV++L++ +T++ GF +T+ G + YS
Sbjct 290 LVNLTNFLVTKHTSALTLQVLGNGKAAVAAGVSVLIFRNPVTVMGIAGFGVTIMGVVLYS 349
Query 147 VSR 149
+R
Sbjct 350 EAR 352
> At2g30460
Length=200
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 78 VVGLGVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLL 137
V+G GV+ + S F + TSP+++ V G++KT L + LL + + G L+
Sbjct 75 VLGFGVLN--VNFSTFLVIGKTSPVTYQVLGHLKTCLVLAFGYLLLKDAFSWRNILGILV 132
Query 138 TLGGSLWYS 146
+ G + YS
Sbjct 133 AVIGMVLYS 141
> At4g32270_1
Length=202
Score = 32.0 bits (71), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 83 VVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWG 124
V+ FVL IF +L S L+ ++ G +K L VG+ +L+G
Sbjct 120 VLAFVLNYCIFLNTTLNSALTQTICGNMKDLFTVGLGWMLFG 161
> At1g76340
Length=372
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 100 SPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGGSLWYSVSRCQ 151
S F+V G V LL V + +++W + T V T G L+ + G + Y S +
Sbjct 277 SATGFTVLGIVNKLLTVVINLMVWDKHSTFVGTLGLLVCMFGGVMYQQSTIK 328
> ECU06g1290
Length=307
Score = 30.8 bits (68), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 37/68 (54%), Gaps = 0/68 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
G ++F+L + F VS T + S+AG VK L+ + A++ L+ + G +++ G
Sbjct 234 GALSFMLLCAEFTLVSETGVVFLSIAGIVKELIIISYAIITKDIALSNLNYVGLTISIAG 293
Query 142 SLWYSVSR 149
L Y+ SR
Sbjct 294 ILIYNFSR 301
> CE23995
Length=332
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 44/88 (50%), Gaps = 5/88 (5%)
Query 38 YNMLNALCLFPIVVLVSEEPKVLWKLAAEGSMMSGTVLAQVVGLGVVTFVLGTSIFWTVS 97
YN + A+ LF +++++ + +W S ++ + G V+ +V G W +
Sbjct 244 YNNILAVLLFLPLIIINGDFGKIWNHFPTWSFWQLLFISGIFGF-VMNYVTG----WQIK 298
Query 98 LTSPLSFSVAGYVKTLLQVGVAMLLWGE 125
TSPL+ +++ K+ Q +A+ L+ E
Sbjct 299 ATSPLTHNISATAKSASQTVIAVFLYSE 326
> YJL193w
Length=402
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 34/74 (45%), Gaps = 2/74 (2%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
G F+ F + S L++S+A +K + V+ + G R+T ++ G +L G
Sbjct 323 GTFHFIQAMITFHLLGEVSTLTYSIANLMKRFAIIAVSWVFIGRRITWLQVFGLVLNTLG 382
Query 142 SLWYSVSRCQANAR 155
Y RC + ++
Sbjct 383 LFLY--ERCTSQSK 394
> 7297822
Length=394
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 11 REAETRDSPPPPLSPVERPRAEPLCMFYNMLNALCLFPIVVLVSE 55
R+A +RD+PP P + P E + ++L A L PI VS+
Sbjct 25 RDAFSRDAPPMPARNHDHPVFEDIRTILSVLKASGLMPIYEQVSD 69
> YML038c
Length=442
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 42/87 (48%), Gaps = 3/87 (3%)
Query 68 SMMSGTVLAQVVGLGVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERL 127
S++ G VL + G V F+L F + T L+ S+ G VK LL V +++ ERL
Sbjct 297 SIVRGIVLLILPGFAV--FLLTICEFSILEQTPVLTVSIVGIVKELLTVIFGIIILSERL 354
Query 128 T-LVETCGFLLTLGGSLWYSVSRCQAN 153
+ G L+ + +Y+ R + +
Sbjct 355 SGFYNWLGMLIIMADVCYYNYFRYKQD 381
> Hs19923531
Length=351
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 34/68 (50%), Gaps = 0/68 (0%)
Query 82 GVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGG 141
G+ F +G + TSPL+ +V+G K Q +A+L + E + + ++ LGG
Sbjct 261 GLFGFAIGYVTGLQIKFTSPLTHNVSGTAKACAQTVLAVLYYEETKSFLWWTSNMMVLGG 320
Query 142 SLWYSVSR 149
S Y+ R
Sbjct 321 SSAYTWVR 328
> YFR032c
Length=289
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 13/21 (61%), Gaps = 0/21 (0%)
Query 9 GLREAETRDSPPPPLSPVERP 29
G AET D PPPP S +RP
Sbjct 264 GQDNAETNDVPPPPASSSDRP 284
> At5g19980
Length=341
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 49/113 (43%), Gaps = 2/113 (1%)
Query 38 YNMLNALCLFPIV-VLVSEEPKVLWKLAAE-GSMMSGTVLAQVVGLGVVTFVLGTSIFWT 95
YN L +L + P+ L E +V L+ G++ + V V F++ F
Sbjct 193 YNNLLSLMIAPVFWFLTGEFTEVFAALSENRGNLFEPYAFSSVAASCVFGFLISYFGFAA 252
Query 96 VSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETCGFLLTLGGSLWYSVS 148
+ S +F+V G V L V + +L+W + T V L T+ G + Y S
Sbjct 253 RNAISATAFTVTGVVNKFLTVVINVLIWDKHATPVGLVCLLFTICGGVGYQQS 305
> At5g42420
Length=350
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 41/85 (48%), Gaps = 1/85 (1%)
Query 62 KLAAEGSMMSGTVLAQVVGLGVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAML 121
K + +M SG L ++ G+ F S + + S +SF V G++KT+ + + L
Sbjct 217 KFIMKYNMSSGCFLFILLSCGLAVFC-NISQYLCIGRFSAVSFQVIGHMKTVCILTLGWL 275
Query 122 LWGERLTLVETCGFLLTLGGSLWYS 146
L+ +T G ++ + G + YS
Sbjct 276 LFDSAMTFKNVAGMIVAIVGMVIYS 300
> SPBC83.11
Length=434
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 17/76 (22%), Positives = 36/76 (47%), Gaps = 0/76 (0%)
Query 74 VLAQVVGLGVVTFVLGTSIFWTVSLTSPLSFSVAGYVKTLLQVGVAMLLWGERLTLVETC 133
V ++ G+ F F +S+ SP+++S+A +K + + V+++ + + +
Sbjct 224 VFLNLIYNGLSHFFQNILAFTLLSIISPVAYSIASLIKRIFVIVVSIIWFQQATNFTQGS 283
Query 134 GFLLTLGGSLWYSVSR 149
G LT G Y S+
Sbjct 284 GIFLTAIGLWLYDRSK 299
> At4g39390
Length=342
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 100 SPLSFSVAGYVKTLLQVGVAMLLWG-ERLTLVETCGFLLTLGGSLWY 145
+ +SF V G++KT+L + + +G E L L G L+ + G +WY
Sbjct 261 TAVSFQVLGHMKTILVLVLGFTFFGKEGLNLQVVLGMLIAILGMIWY 307
> At1g56140
Length=2062
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 57 PKVLWKLAAEGSMMSGTVLAQVVGLGVVTFVLGTSIF 93
P V + ++G M+GT++ +VG+G+++ + G IF
Sbjct 1649 PTVGNRPPSKGKSMTGTIVGVIVGVGLLSIISGVVIF 1685
Lambda K H
0.321 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40