bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1552_orf1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
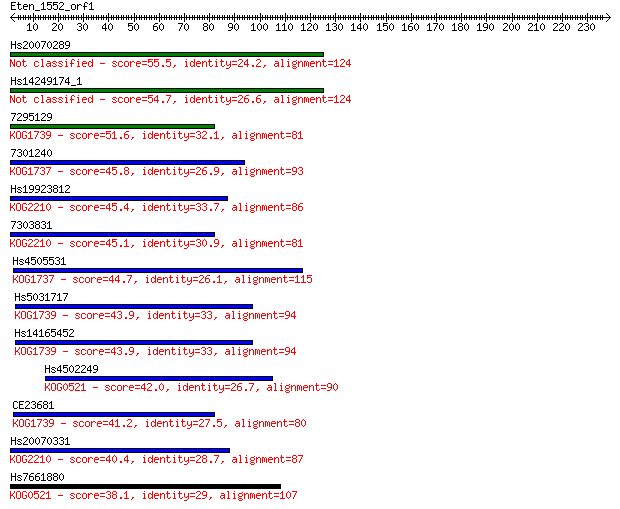
Hs20070289 55.5 1e-07
Hs14249174_1 54.7 2e-07
7295129 51.6 2e-06
7301240 45.8 7e-05
Hs19923812 45.4 1e-04
7303831 45.1 1e-04
Hs4505531 44.7 2e-04
Hs5031717 43.9 3e-04
Hs14165452 43.9 3e-04
Hs4502249 42.0 0.001
CE23681 41.2 0.002
Hs20070331 40.4 0.003
Hs7661880 38.1 0.014
> Hs20070289
Length=300
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/127 (23%), Positives = 58/127 (45%), Gaps = 4/127 (3%)
Query 1 LQGWLQKWTNLLSSWRPRYFFVYAGFFKYSTTKSSP---PKEMFLLNQCRVRLCPHDPVR 57
++G L KWTN L+ W+PR+F + G Y ++ K + C +++ D R
Sbjct 1 MEGVLYKWTNYLTGWQPRWFVLDNGILSYYDSQDDVCKGSKGSIKMAVCEIKVHSADNTR 60
Query 58 FEVDVVDQQTLYLRAESQEEKQMWYAAFKQGQRAALSGANRANRSAAGGGSQRDRQQQQQ 117
E+ + +Q Y++A + E+Q W A +A L+ S+ + + +
Sbjct 61 MELIIPGEQHFYMKAVNAAERQRWLVALG-SSKACLTDTRTKKEKEISETSESLKTKMSE 119
Query 118 LQMHRRL 124
L+++ L
Sbjct 120 LRLYCDL 126
> Hs14249174_1
Length=326
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/130 (25%), Positives = 59/130 (45%), Gaps = 10/130 (7%)
Query 1 LQGWLQKWTNLLSSWRPRYFFVYAGFFKYSTTKSSPP------KEMFLLNQCRVRLCPHD 54
++G L KWTN LS W+PR+F + G Y SP K + C +++ D
Sbjct 1 MEGVLYKWTNYLSGWQPRWFLLCGGILSYY---DSPEDAWKGCKGSIQMAVCEIQVHSVD 57
Query 55 PVRFEVDVVDQQTLYLRAESQEEKQMWYAAFKQGQRAALSGANRANRSAAGGGSQRDRQQ 114
R ++ + +Q YL+A S E+Q W A +A L+ + ++ + +
Sbjct 58 NTRMDLIIPGEQYFYLKARSVAERQRWLVALGSA-KACLTDSRTQKEKEFAENTENLKTK 116
Query 115 QQQLQMHRRL 124
+L+++ L
Sbjct 117 MSELRLYCDL 126
> 7295129
Length=601
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 43/84 (51%), Gaps = 3/84 (3%)
Query 1 LQGWLQKWTNLLSSWRPRYFFVYAGFFKYSTTKSSPP---KEMFLLNQCRVRLCPHDPVR 57
L+G+L KWTN + W+PRY + G Y ++S + L + ++ D +R
Sbjct 42 LRGYLSKWTNYIYGWQPRYIVLKDGTLSYYKSESESDFGCRGAISLTKATIKAHESDELR 101
Query 58 FEVDVVDQQTLYLRAESQEEKQMW 81
F+V V + LRAE+ E++ W
Sbjct 102 FDVVVNNLNNWCLRAETSEDRMHW 125
> 7301240
Length=784
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 46/100 (46%), Gaps = 13/100 (13%)
Query 1 LQGWLQKWTNLLSSWRPRYFFVYAGFFKYSTTKSSPPKEMFLLNQCRVRLCPH------- 53
++GWL KWTN + ++ R+F + G Y +S + + CR + H
Sbjct 17 MKGWLLKWTNYIKGYQRRWFVLSKGVLSYYRNQSE------INHTCRGTISLHGALIHTV 70
Query 54 DPVRFEVDVVDQQTLYLRAESQEEKQMWYAAFKQGQRAAL 93
D F + QT +++A ++ E+Q W A + + A+
Sbjct 71 DSCTFVISNGGTQTFHIKAGTEVERQSWVTALELAKAKAI 110
> Hs19923812
Length=764
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 42/91 (46%), Gaps = 5/91 (5%)
Query 1 LQGWLQKWTNLLSSWRPRYFFV--YAGFFKYST---TKSSPPKEMFLLNQCRVRLCPHDP 55
L+G L K+TNLL W+ RYF + AG +Y +K P+ + L+ V L P
Sbjct 77 LEGVLSKYTNLLQGWQNRYFVLDFEAGILQYFVNEQSKHQKPRGVLSLSGAIVSLSDEAP 136
Query 56 VRFEVDVVDQQTLYLRAESQEEKQMWYAAFK 86
V + + LRA +EKQ W +
Sbjct 137 HMLVVYSANGEMFKLRAADAKEKQFWVTQLR 167
> 7303831
Length=764
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 42/87 (48%), Gaps = 7/87 (8%)
Query 1 LQGWLQKWTNLLSSWRPRYFFV--YAGFFKYSTTKSSPPKEM----FLLNQCRVRLCPHD 54
L+G L KWTN++ W+ R+F + AG Y T+K K + L + + +
Sbjct 24 LEGTLSKWTNVMKGWQYRFFVLDENAGLLSYYTSKDKMIKGVRRGCVRLKDALIGIDDQE 83
Query 55 PVRFEVDVVDQQTLYLRAESQEEKQMW 81
F + VD +T + +A EE++ W
Sbjct 84 DNTFTI-TVDHKTFHFQARHNEEREQW 109
> Hs4505531
Length=807
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 30/117 (25%), Positives = 52/117 (44%), Gaps = 2/117 (1%)
Query 2 QGWLQKWTNLLSSWRPRYFFVYAGFFKYSTTKSSPPKEMF-LLNQCRVRLCPHDPVRFEV 60
+GWL KWTN + ++ R+F + G Y +K+ +N + D F +
Sbjct 92 EGWLFKWTNYIKGYQRRWFVLSNGLLSYYRSKAEMRHTCRGTINLATANITVEDSCNFII 151
Query 61 DVVDQQTLYLRAESQEEKQMWYAAFKQGQRAALSGANRANRSAAGGG-SQRDRQQQQ 116
QT +L+A S+ E+Q W A + + A+ ++ S SQ D+ + Q
Sbjct 152 SNGGAQTYHLKASSEVERQRWVTALELAKAKAVKMLAESDESGDEESVSQTDKTELQ 208
> Hs5031717
Length=624
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 45/103 (43%), Gaps = 16/103 (15%)
Query 3 GWLQKWTNLLSSWRPRYFFVYAGFFKYSTTKSSPPKEMFLLNQCRVRLC-------PH-- 53
G L KWTN + W+ R+ + Y KS E CR +C PH
Sbjct 28 GVLSKWTNYIHGWQDRWVVLKNNALSY--YKSEDETEY----GCRGSICLSKAVITPHDF 81
Query 54 DPVRFEVDVVDQQTLYLRAESQEEKQMWYAAFKQGQRAALSGA 96
D RF++ V D YLRA+ + +Q W A +Q + + G+
Sbjct 82 DECRFDISVND-SVWYLRAQDPDHRQQWIDAIEQHKTESGYGS 123
> Hs14165452
Length=598
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 45/103 (43%), Gaps = 16/103 (15%)
Query 3 GWLQKWTNLLSSWRPRYFFVYAGFFKYSTTKSSPPKEMFLLNQCRVRLC-------PH-- 53
G L KWTN + W+ R+ + Y KS E CR +C PH
Sbjct 28 GVLSKWTNYIHGWQDRWVVLKNNALSY--YKSEDETEY----GCRGSICLSKAVITPHDF 81
Query 54 DPVRFEVDVVDQQTLYLRAESQEEKQMWYAAFKQGQRAALSGA 96
D RF++ V D YLRA+ + +Q W A +Q + + G+
Sbjct 82 DECRFDISVND-SVWYLRAQDPDHRQQWIDAIEQHKTESGYGS 123
> Hs4502249
Length=1006
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/91 (26%), Positives = 47/91 (51%), Gaps = 3/91 (3%)
Query 15 WRPRYFFVYAGFFKYST-TKSSPPKEMFLLNQCRVRLCPHDPVRFEVDVVDQQTLYLRAE 73
W+ R V GF S T + PP ++ LL C+V+ P + F++ + +T + +AE
Sbjct 323 WQKRKCSVKNGFLTISHGTANRPPAKLNLLT-CQVKTNPEEKKCFDL-ISHDRTYHFQAE 380
Query 74 SQEEKQMWYAAFKQGQRAALSGANRANRSAA 104
++E Q+W + + + AL+ A + + +
Sbjct 381 DEQECQIWMSVLQNSKEEALNNAFKGDDNTG 411
> CE23681
Length=573
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 38/83 (45%), Gaps = 3/83 (3%)
Query 2 QGWLQKWTNLLSSWRPRYFFVYAGFFKYSTT---KSSPPKEMFLLNQCRVRLCPHDPVRF 58
G ++KWTN ++ W+ RYF + G Y + KS + L + D F
Sbjct 10 NGVMKKWTNYVNGWQDRYFEITEGNLVYYLSKAEKSHGCRGSIFLKSAIIAAHEFDENEF 69
Query 59 EVDVVDQQTLYLRAESQEEKQMW 81
+ + + YL+AE+ + K +W
Sbjct 70 SISMGENVVWYLKAENSQSKLLW 92
> Hs20070331
Length=736
Score = 40.4 bits (93), Expect = 0.003, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 45/93 (48%), Gaps = 7/93 (7%)
Query 1 LQGWLQKWTNLLSSWRPRYFFV--YAGFFKYSTTKSS----PPKEMFLLNQCRVRLCPHD 54
++G L KWTN++ W+ R+F + AG Y T+K + L + + D
Sbjct 5 MEGPLSKWTNVMKGWQYRWFVLDYNAGLLSYYTSKDKMMRGSRRGCVRLRGAVIGIDDED 64
Query 55 PVRFEVDVVDQQTLYLRAESQEEKQMWYAAFKQ 87
F + VDQ+T + +A +E++ W A ++
Sbjct 65 DSTFTI-TVDQKTFHFQARDADEREKWIHALEE 96
> Hs7661880
Length=740
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 53/110 (48%), Gaps = 4/110 (3%)
Query 1 LQGWL-QKWTNLLSSWRPRYFFVYAGFFKYSTTKSSPPKEMFL-LNQCRVRLCPHDPVRF 58
++G L ++ +N +W R+F + + Y P + L C V+LCP RF
Sbjct 268 MEGHLFKRASNAFKTWSRRWFTIQSNQLVYQKKYKDPVTVVVDDLRLCTVKLCPDSERRF 327
Query 59 EVDVVDQ-QTLYLRAESQEEKQMWYAAFKQGQRAALSGANRANRSAAGGG 107
+VV ++ L+A+S+ Q+W +A + +A S A R + S G G
Sbjct 328 CFEVVSTSKSCLLQADSERLLQLWVSAVQSSIASAFSQA-RLDDSPRGPG 376
Lambda K H
0.311 0.120 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4740636838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40