bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1610_orf4
Length=191
Score E
Sequences producing significant alignments: (Bits) Value
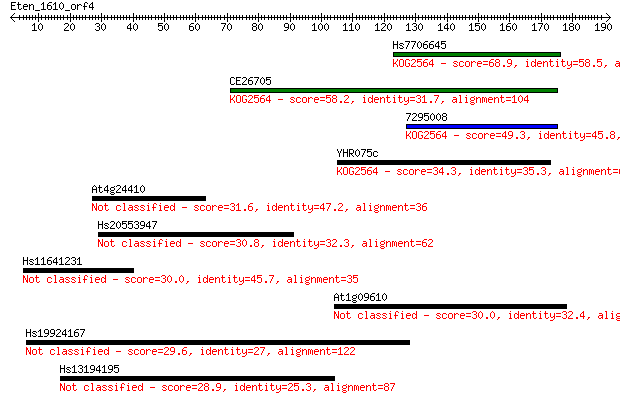
Hs7706645 68.9 6e-12
CE26705 58.2 1e-08
7295008 49.3 5e-06
YHR075c 34.3 0.14
At4g24410 31.6 1.1
Hs20553947 30.8 1.6
Hs11641231 30.0 2.8
At1g09610 30.0 3.1
Hs19924167 29.6 3.8
Hs13194195 28.9 7.0
> Hs7706645
Length=386
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 39/55 (70%), Gaps = 3/55 (5%)
Query 123 PLPTWDEFFEKCEDIRPLESS--DTFRVYSSGSSGPLLFLLHGAGHTSLSWACFT 175
P+P W ++FE ED+ + DTFRVY SGS GP+L LLHG GH++LSWA FT
Sbjct 43 PVP-WSQYFESMEDVEVENETGKDTFRVYKSGSEGPVLLLLHGGGHSALSWAVFT 96
> CE26705
Length=364
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 51/104 (49%), Gaps = 11/104 (10%)
Query 71 DFETEAATEHRPHSRQPRSASSARQRTGRHRDAQTPHSGSSSRRSSCNASCGPLPTWDEF 130
D ++E + PH R RQ R P + +S ++ + LP W +F
Sbjct 11 DLQSETSHVTTPH----RQNDLLRQAVTHGRPPPVPSTSTSGKKREMSE----LP-WSDF 61
Query 131 FEKCEDIRPLESSDTFRVYSSGSSGPLLFLLHGAGHTSLSWACF 174
F++ +D D F VY G+ GP+ +LLHG G++ L+WACF
Sbjct 62 FDEKKDANI--DGDVFNVYIKGNEGPIFYLLHGGGYSGLTWACF 103
> 7295008
Length=402
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 34/51 (66%), Gaps = 4/51 (7%)
Query 127 WDEFFEKCEDIRPLESSDTFRVYSSG---SSGPLLFLLHGAGHTSLSWACF 174
W+EFF + ED+ ++ TFR+Y + GP+L LLHG G+++L+WA F
Sbjct 34 WNEFFAEKEDVT-VDEQRTFRIYRTKQPEKPGPVLLLLHGGGYSALTWAHF 83
> YHR075c
Length=400
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 39/76 (51%), Gaps = 9/76 (11%)
Query 105 TPHSGSSSRRSSCNASCGPLPTWDEFFEKCEDI----RPLESSDTFR----VYSSGSSGP 156
T +GS++ R S LPTW +FF+ E + R L+ + + + S+ +S P
Sbjct 56 TGKTGSTTDRISSKEKSS-LPTWSDFFDNKELVSLPDRDLDVNTYYTLPTSLLSNTTSIP 114
Query 157 LLFLLHGAGHTSLSWA 172
+ HGAG + LS+A
Sbjct 115 IFIFHHGAGSSGLSFA 130
> At4g24410
Length=169
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query 27 GSPSESPHQSRRKSAGCS--MRPVKEERQQAEPEHAPE 62
GSPS P + RRK+ G S +RP++E+R+ E A E
Sbjct 121 GSPSRDPIEIRRKTVGKSRDVRPIQEKREARARERASE 158
> Hs20553947
Length=760
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 29 PSESPHQSRRKSAGCSMRPVKEERQQAEPEHAPEGSASHANLDFETEAATEHRPHSRQPR 88
P E ++R K+ C+ +PVKEE+ Q E AP+ ++ A+L + T + S P
Sbjct 119 PKEKEEKNRDKNKVCTEKPVKEEKDQV-TEMAPKKTSKIASLIPKGSKTTAAKKESLIPS 177
Query 89 SA 90
S+
Sbjct 178 SS 179
> Hs11641231
Length=448
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 5 PPDIKELPPITRGRSSNFPDRVGSPSESPHQSRRK 39
PP+ KE T G S+NFP GS + SP Q K
Sbjct 142 PPNTKECVGATSGASANFPGTSGSSTLSPFQHAHK 176
> At1g09610
Length=282
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 31/79 (39%), Gaps = 8/79 (10%)
Query 104 QTPHSGSSSRRSSCNASCGPLP---TWDEFFEKCEDIRPLESSDTFRVYS--SGSSGPLL 158
QTP S+R C+ +C LP I P ++ V S G P
Sbjct 39 QTPQETRSTR---CSGACNKLPRSLAQALIHYSTSVITPQQTLKEIAVSSRVLGKKSPCN 95
Query 159 FLLHGAGHTSLSWACFTVG 177
FL+ G GH SL W+ G
Sbjct 96 FLVFGLGHDSLMWSSLNYG 114
> Hs19924167
Length=531
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 46/125 (36%), Gaps = 29/125 (23%)
Query 6 PDIKELPPITRGRSSNFPDRVGSPSESPHQSRRKSAGCSMRP---VKEERQQAEPEHAPE 62
P ++ LPP + D V + + R K+ G +P + E PE PE
Sbjct 77 PLLQPLPPSKAAEELHRVDLVLPEDTTEYFVRTKAGGVCFKPGTKMLERPPPGRPEEKPE 136
Query 63 GSASHANLDFETEAATEHRPHSRQPRSASSARQRTGRHRDAQTPHSGSSSRRSSCNASCG 122
G+ ++ RP PR SAR+RTG G +RR C
Sbjct 137 GA----------NGSSARRP----PRYLLSARERTG----------GRGARRKWVECVC- 171
Query 123 PLPTW 127
LP W
Sbjct 172 -LPGW 175
> Hs13194195
Length=501
Score = 28.9 bits (63), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 40/88 (45%), Gaps = 14/88 (15%)
Query 17 GRSSNFPDRVGSPSESPHQSRRKSAGCSMRPVKEERQQAEPEHAPEGSASHANLDFETEA 76
G+S + +G P P Q+ +RP ++ R+Q E ++ P+G A A+ + +
Sbjct 304 GKSVHVLRHIGGPESEPPQA--------LRP-RDRRRQEEIDYRPDGGAGDADFHYRGQM 354
Query 77 A-TEHRPHSRQPRSASSARQRTGRHRDA 103
TE P+++ R+ R RD
Sbjct 355 GPTEQGPYAK----TYEGRREILRERDV 378
Lambda K H
0.314 0.127 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3202455494
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40