bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1639_orf2
Length=471
Score E
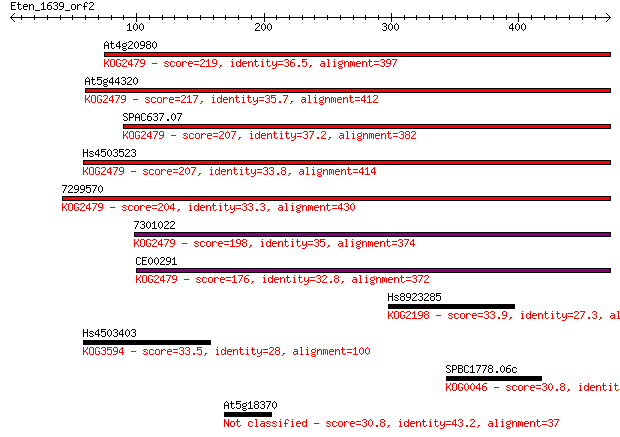
Sequences producing significant alignments: (Bits) Value
At4g20980 219 1e-56
At5g44320 217 5e-56
SPAC637.07 207 3e-53
Hs4503523 207 6e-53
7299570 204 4e-52
7301022 198 2e-50
CE00291 176 1e-43
Hs8923285 33.9 0.86
Hs4503403 33.5 0.95
SPBC1778.06c 30.8 6.1
At5g18370 30.8 7.6
> At4g20980
Length=591
Score = 219 bits (557), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 145/413 (35%), Positives = 225/413 (54%), Gaps = 50/413 (12%)
Query 75 QKKQQQQQQARYARIHARHR-----------AF-SEWSIQPTNDWQVLAELPLQQLHK-- 120
+K+ ++++AR R++ +R AF S IQP +W +L ++P K
Sbjct 149 KKRDAEKERARRDRLYNNNRNNIHHQRREAAAFKSSVDIQP--EWNMLEQIPFSTFSKLS 206
Query 121 QQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDEVIRDY 180
++ P EDL G L Y+R D+I+ K ++ N ++ VTT DD VIR
Sbjct 207 YTVQEP----EDLLLCGGLEYYNRLFDRITPKNERRLERFK-NRNFFKVTTSDDPVIR-- 259
Query 181 MLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLTVNETS 240
L + V ATD +LA LM AP+S YSW + + ++ K+ DK +GS +DLL+V+ETS
Sbjct 260 RLAKEDKATVFATDAILAALMCAPRSVYSWDIVIQRVGNKLFFDKRDGSQLDLLSVHETS 319
Query 241 IEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYPPA-PFVESGDTPATIA 299
EP + ++++ +N +LG EA INQN QQ+LV + A PF G+ A++A
Sbjct 320 QEP-LPESKDDINSAHSLGVEAAYINQNFSQQVLVRDGKKETFDEANPFANEGEEIASVA 378
Query 300 YRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGGYIYTC 359
YRYR + + ++++ R E+ + ADL R ++
Sbjct 379 YRYRRWKLDDN---------MHLVARCEL-----QSVADLNNQR----------SFLTLN 414
Query 360 ALNEIEPK-GQKNWRTLLESQRGALLATEVRNNACKLRKFVTSAMIAGCDELRLGFVARK 418
ALNE +PK +WR LE+QRGA+LATE++NN KL K+ A++A D +++GFV+R
Sbjct 415 ALNEFDPKYSGVDWRQKLETQRGAVLATELKNNGNKLAKWTAQALLANADMMKIGFVSRV 474
Query 419 TPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
P D+ H+IL+V Y+ +D QI L N WGIV++++DL +G+YVL
Sbjct 475 HPRDHFNHVILSVLGYKPKDFAGQINLNTSNMWGIVKSIVDLCMKLSEGKYVL 527
> At5g44320
Length=588
Score = 217 bits (552), Expect = 5e-56, Method: Compositional matrix adjust.
Identities = 147/418 (35%), Positives = 220/418 (52%), Gaps = 39/418 (9%)
Query 60 KDEEELKLGKKAKSDQKKQQQQQQARYARIHARHR---AF-SEWSIQPTNDWQVLAELPL 115
+DEE ++A+ D+ ++ + IH + R AF S IQP +W +L ++P
Sbjct 138 RDEEVEAKKREAEKDRARRDRLYNNNRNNIHQQRREAAAFKSSVDIQP--EWNMLEQIPF 195
Query 116 QQLHKQQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDE 175
K EDL G L Y R D+I+ K ++ + + VTT DD
Sbjct 196 STFSKLSFTVSEP--EDLLLCGGLESYDRSFDRITPKADRRLERFKNRSFK--VTTSDDL 251
Query 176 VIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLT 235
VIR L + V ATD +LA LM AP+S YSW L + ++ K+ DK +GS +DLL+
Sbjct 252 VIR--RLAKEDKATVFATDAILAALMCAPRSVYSWDLVIQRVGNKLFFDKRDGSPLDLLS 309
Query 236 VNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVP-GEVAAEYPPAPFVESGDT 294
V+ETS EP + + ++ +N +LG EA INQN QQ+LV G+ P P V G+
Sbjct 310 VHETSQEP-LPEGKDDINSAHSLGLEAAYINQNFAQQVLVKNGKRETFDEPIPNVNEGEE 368
Query 295 PATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGG 354
A+IAYRYR + + + ++ R E+ + DL R
Sbjct 369 NASIAYRYRRWKLDDS---------MYLVARCEL-----QSTVDLNNQR----------S 404
Query 355 YIYTCALNEIEPK-GQKNWRTLLESQRGALLATEVRNNACKLRKFVTSAMIAGCDELRLG 413
++ ALNE +PK +WR LE+QRGA+LA E++NN KL K+ A++A D +++G
Sbjct 405 FLTLNALNEFDPKYSGVDWRQKLETQRGAVLANELKNNGNKLAKWTAQALLANADMMKIG 464
Query 414 FVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
FV+R P D+ H+IL+V Y+ +D QI L N WGIV++++DL +G+YVL
Sbjct 465 FVSRVHPRDHFNHVILSVLGYKPKDFAGQINLNTNNMWGIVKSIVDLCMKLSEGKYVL 522
> SPAC637.07
Length=567
Score = 207 bits (528), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 142/397 (35%), Positives = 203/397 (51%), Gaps = 43/397 (10%)
Query 90 HARHRAFSEWSIQPTNDWQVLAELPLQQLHKQQIETPR-VTVEDLEWRGTLGVYSRQADK 148
+ +H+ S+ +DWQ+L E+ L K + VTV+ G + Y + DK
Sbjct 166 YDKHQRLRNASVTVGDDWQLLDEVEFSHLSKLNLAAAAPVTVDSY---GYIYPYDKSFDK 222
Query 149 ISSKLPVPMQNLSSNFYYYWVTTQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKY 208
I K P+Q L + +Y TT +D VI+ LN + + TD +L+ LM + +S Y
Sbjct 223 IHVKSEKPLQAL--DRVHYNPTTTEDPVIQKLALNSDANIFI--TDSILSLLMCSTRSVY 278
Query 209 SWHLYLTKIDGKIIIDKANGSIVDLLTVNETSIEPPMQDAENR--LNRPAALGFEAVRIN 266
W + +T GK+ DK G D LTVNE + + PM DA+NR +N P AL EA IN
Sbjct 279 PWDIVITHQSGKLFFDKREGGPFDYLTVNENAYDSPM-DADNREGVNSPGALSVEATYIN 337
Query 267 QNL-RQQLLVPGEVAAEYP-PAPFVESGDTPATIA---YRYRLFTIPPRAGPNASRRPIN 321
QN Q L E + P P PF S + +A Y YR + +P+
Sbjct 338 QNFCVQALRETEEEKYKLPHPNPFYNSKEESEPLAAHGYIYRDVDL----SLETDEKPVK 393
Query 322 ILTRAEV--YAKSPMARADLKEAREGDNFIVDGGGYIYTCALNEIEPK-----GQKNWRT 374
++ R EV Y K+P D++ YI ALNE +PK G +WR+
Sbjct 394 LMVRTEVDGYVKNPAN--DVQ--------------YISIKALNEYDPKFTNVTGSVDWRS 437
Query 375 LLESQRGALLATEVRNNACKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNY 434
LESQRGA+ ATE++NN+CKL ++ A++AG D +++GFV+R D + H IL V Y
Sbjct 438 KLESQRGAVFATEMKNNSCKLARWTVEALLAGVDSMKVGFVSRSNARDAQHHGILGVVAY 497
Query 435 QTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
+ DL Q+ L N WGIVR + D+ PDG+YVL
Sbjct 498 KPADLASQMNLSLSNGWGIVRTIADVCLKMPDGKYVL 534
> Hs4503523
Length=548
Score = 207 bits (526), Expect = 6e-53, Method: Compositional matrix adjust.
Identities = 140/427 (32%), Positives = 220/427 (51%), Gaps = 58/427 (13%)
Query 58 KTKDEEELKLGKKAKSD----QKKQQQQQQARYARIHARHRAFSEWSIQPTNDWQVLAEL 113
K K+ E ++L KK + QK Q+ Q+ R + + R +DW+V E+
Sbjct 127 KQKERERIRLQKKFQKQFGVRQKWDQKSQKPRDSSVEVR------------SDWEVKEEM 174
Query 114 PLQQLHKQ---QIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVT 170
QL K ++ P +D+E G L Y + D+I+++ P++++ F+ VT
Sbjct 175 DFPQLMKMRYLEVSEP----QDIECCGALEYYDKAFDRITTRSEKPLRSIKRIFHT--VT 228
Query 171 TQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSI 230
T DD VIR Q +V ATD +LA LM+ +S YSW + + ++ K+ DK + S
Sbjct 229 TTDDPVIRKL---AKTQGNVFATDAILATLMSCTRSVYSWDIVVQRVGSKLFFDKRDNSD 285
Query 231 VDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYP-PAPFV 289
DLLTV+ET+ EPP QD N N P L EA IN N QQ L G+ +P P PFV
Sbjct 286 FDLLTVSETANEPP-QDEGNSFNSPRNLAMEATYINHNFSQQCLRMGKERYNFPNPNPFV 344
Query 290 ESG---DTPATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGD 346
E + A++AYRYR + + I+++ R E A ++
Sbjct 345 EDDMDKNEIASVAYRYRRWKLGD---------DIDLIVRCEHDGVMTGANGEV------- 388
Query 347 NFIVDGGGYIYTCALNEIEPKGQK--NWRTLLESQRGALLATEVRNNACKLRKFVTSAMI 404
+I LNE + + +WR L+SQRGA++ATE++NN+ KL ++ A++
Sbjct 389 -------SFINIKTLNEWDSRHCNGVDWRQKLDSQRGAVIATELKNNSYKLARWTCCALL 441
Query 405 AGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDK 464
AG + L+LG+V+R D+ +H+IL ++ + QI L ENAWGI+R V+D+
Sbjct 442 AGSEYLKLGYVSRYHVKDSSRHVILGTQQFKPNEFASQINLSVENAWGILRCVIDICMKL 501
Query 465 PDGRYVL 471
+G+Y++
Sbjct 502 EEGKYLI 508
> 7299570
Length=551
Score = 204 bits (519), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 143/439 (32%), Positives = 223/439 (50%), Gaps = 42/439 (9%)
Query 42 VSTRGKLKQTT---AAFNEKTKDEEELKLGKKAKSDQKKQQQQQQARYARIHARHRAFSE 98
V +RG+ + T A+ T K K + Q R+ R +A R E
Sbjct 107 VRSRGRTGRGTPNIASLGGSTAGGATASTTKYGKGRNTRNTQNMGRRFGR-NAPTR-IRE 164
Query 99 WSIQPTNDWQVLAELPLQQLHKQQIETPRVT-VEDLEWRGTLGVYSRQADKISSKLPVPM 157
S+ +DW + E+ +L K + P + +D+ G L Y + D+++ + P+
Sbjct 165 SSVMVQSDWVSIEEIDFPRLLK--LALPNIKDGKDIATCGWLEFYDKLYDRVNLRNEKPL 222
Query 158 QNLSSNFYYYWVTTQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKI 217
Q + + VTT DD VIR +V ATD++L+ +M +S YSW + + K+
Sbjct 223 QKMDR--VVHTVTTTDDPVIRRL---SRTMGNVFATDEILSTIMCCTRSNYSWDVVVEKL 277
Query 218 DGKIIIDKANGSIVDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPG 277
K+ +DK DLLTVNETS+EPPM D E +N +L EA IN N QQ+L G
Sbjct 278 GTKVFLDKRYNDQFDLLTVNETSVEPPM-DEEGSINSAHSLAMEATLINHNFSQQVLRIG 336
Query 278 EVAAEY---PPAPFVESGDTPATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPM 334
+ + P PF E G A+I YRYR + + +L +
Sbjct 337 DQEPRFMFEEPNPFEEPGVDLASIGYRYRQWDLGNDV----------VL----------I 376
Query 335 ARADLKEAREGDNFIVDGGGYIYTCALNEIEPK--GQKNWRTLLESQRGALLATEVRNNA 392
AR +G N V ++ ALNE + K WR L++QRGA+LA+E+RNNA
Sbjct 377 ARCKHNGVIQGPNGDVQ---FLSIKALNEWDSKVTNSVEWRQKLDTQRGAVLASELRNNA 433
Query 393 CKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWG 452
CKL ++ A++AG D+L+LG+V+R P D+ +H+IL ++ ++ QI L +NAWG
Sbjct 434 CKLARWTVEAVLAGSDQLKLGYVSRMNPRDHLRHVILGTQQFKPQEFATQINLNMDNAWG 493
Query 453 IVRAVLDLLEDKPDGRYVL 471
++R ++DL+ +PDG+Y++
Sbjct 494 VLRCLIDLVMRQPDGKYLI 512
> 7301022
Length=560
Score = 198 bits (504), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 131/385 (34%), Positives = 202/385 (52%), Gaps = 45/385 (11%)
Query 98 EWSIQPTNDWQVLAELPLQQLHKQQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPM 157
E S+ DW + E+ +L K + + V D+ GTL Y + D+I+ K P+
Sbjct 165 ESSVAVRADWASIEEMDFPRLIKLSLPNIKEGV-DIVTCGTLEYYDKTYDRINVKNEKPL 223
Query 158 QNLSSNFYYYWVTTQDDEVIRDYMLNGSKQV-DVAATDQVLACLMAAPQSKYSWHLYLTK 216
Q + + VTT DD VIR SK V +V ATD +LA +M + +S YSW + + K
Sbjct 224 QKIDR--IVHTVTTTDDPVIRRL----SKTVGNVFATDAILATIMCSTRSNYSWDIVIEK 277
Query 217 IDGKIIIDKANGSIVDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVP 276
+ K+ +DK + + DLLTVNE+S+EPP D ++ N P L EA IN N QQ+L
Sbjct 278 VGDKVFMDKRDHTEFDLLTVNESSVEPPTDD-DSSCNSPRNLAIEATFINHNFSQQVLKT 336
Query 277 GEVAAEYP---PAPFVESGDT--PATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAK 331
G+ +Y PF+ + A++ YRY+ + + I ++ R E
Sbjct 337 GDQEPKYKFEESNPFISEDEDIQVASVGYRYKKWELG---------SDIVLVARCE---- 383
Query 332 SPMARADLKEAREGDNFIVDGGG---YIYTCALNEIEPK--GQKNWRTLLESQRGALLAT 386
D + G ++ ALNE + K WR L++QRGA+LA
Sbjct 384 -------------HDGVLQTPSGEPQFMTIKALNEWDSKLANGVEWRQKLDTQRGAVLAN 430
Query 387 EVRNNACKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLR 446
E+RNNACKL K+ A++AG D+L+LG+V+R P D+ +H+IL ++ + QI L
Sbjct 431 ELRNNACKLAKWTVQAVLAGSDQLKLGYVSRINPRDHSRHVILGTQQFKPHEFATQINLS 490
Query 447 PENAWGIVRAVLDLLEDKPDGRYVL 471
+NAWGI+R ++DL+ + DG+Y++
Sbjct 491 MDNAWGILRCIIDLVMKQKDGKYLI 515
> CE00291
Length=570
Score = 176 bits (446), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 122/387 (31%), Positives = 193/387 (49%), Gaps = 44/387 (11%)
Query 100 SIQPTNDWQVLAELPLQQLHKQQIETPRV----TVEDLEWRGTLGVYSRQADKISSKLPV 155
S+Q +W VL E+ L K + P + + D ++ G+L Y + D++S K +
Sbjct 178 SVQVRPEWVVLEEMNLSAFSK--LALPNIPGGDDIGDHQY-GSLQYYDKTIDRVSVKNSI 234
Query 156 PMQNLSSNFYYYWVTTQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLT 215
P+Q + FY VTT +D VI++ G+ +V TD +LA LM AP+S YSW +
Sbjct 235 PLQRCAGVFYN--VTTTEDPVIQELAQGGAG--NVFGTDIILATLMTAPRSVYSWDIVAY 290
Query 216 KIDGKIIIDKANG----SIVDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQ 271
++ K+ DK N + V+ LTV+ETS EPP D N +N L EA INQN R+
Sbjct 291 RVGDKLFFDKRNTRDILNPVETLTVSETSAEPPSFDG-NGINNAKDLATEAFYINQNFRR 349
Query 272 QLLVPGEVAAEYPPA--PFVESGDTPATIAYRYRLFTIPPRAGPNASRRPINILTRAEVY 329
Q++ + + A PF + + AY+YR + + G +P+ ++ R E+
Sbjct 350 QVVKRNDAGFTFKNARAPFEDEETGESGTAYKYRKWNL----GNGVDGKPVELVCRTEL- 404
Query 330 AKSPMARADLKEAREGDNFIVDGGGYIYTC---ALNEIEP--KGQKNWRTLLESQRGALL 384
D I G T A NE + G +WRT L+ Q+GA++
Sbjct 405 ----------------DGVIHGLGNETQTLTIKAFNEWDSTQSGGVDWRTKLDVQKGAVM 448
Query 385 ATEVRNNACKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIG 444
ATE++NN+ K+ K+ A++AG D ++LG+V+R + H IL + + I
Sbjct 449 ATEIKNNSAKVAKWTLQALLAGSDTMKLGYVSRNNARSTQNHSILLTQYVKPTEFASNIA 508
Query 445 LRPENAWGIVRAVLDLLEDKPDGRYVL 471
L +N WGI+R V+D + G+Y+L
Sbjct 509 LNMDNCWGILRCVIDSCMKQKPGKYLL 535
> Hs8923285
Length=531
Score = 33.9 bits (76), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 44/100 (44%), Gaps = 3/100 (3%)
Query 298 IAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGGYIY 357
I + R+ P +G R+ I++ + + L+ A G + +GG +Y
Sbjct 23 ILFYDRILCDVPCSGDGTMRKNIDVWKKWTTLNSLQLHGLQLRIATRGAEQLAEGGRVVY 82
Query 358 -TCALNEIEPKGQKNWRTLLESQRGALLATEVRNNACKLR 396
TC+LN IE + +LLE GAL +V N L+
Sbjct 83 STCSLNPIE--DEAVIASLLEKSEGALELADVSNELPGLK 120
> Hs4503403
Length=1117
Score = 33.5 bits (75), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 50/101 (49%), Gaps = 5/101 (4%)
Query 58 KTKDEEELKLGKKAKSDQKKQQQQQQARYARIH-ARHRAFSEWSIQPTNDWQVLAELPLQ 116
K K E+ LG+K +++ +Q+Q+ A ++ A H + + N + + P+
Sbjct 831 KFKTLAEVCLGQKIDINKEIEQRQKPATETSMNTASHSLCEQTMVNSENTYSSGSSFPVP 890
Query 117 QLHKQQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPM 157
+ Q+ +VT E + R V SRQA K+++ LP PM
Sbjct 891 K-SLQEANAEKVTQEIVTERS---VSSRQAQKVATPLPDPM 927
> SPBC1778.06c
Length=614
Score = 30.8 bits (68), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 41/79 (51%), Gaps = 6/79 (7%)
Query 343 REGDNFIVD-GGGYIYTCALNEIEPKGQKNWRTLLESQRGALLATEVRNNACKL--RKFV 399
R NF D G YT LN++ P + R L++ A +V NA KL RK++
Sbjct 282 RTVSNFSKDVSDGENYTVLLNQLAP--ELCSRAPLQTTDVLQRAEQVLQNAEKLDCRKYL 339
Query 400 T-SAMIAGCDELRLGFVAR 417
T +AM+AG +L L FVA
Sbjct 340 TPTAMVAGNPKLNLAFVAH 358
> At5g18370
Length=1210
Score = 30.8 bits (68), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query 169 VTTQDDEVIRDYMLNGSKQVDVAATDQVLA--CLMAAPQ 205
+TTQD ++R + +N +VD+ ATD+ L CL A Q
Sbjct 385 ITTQDQRLLRAHEINHVYKVDLPATDEALQIFCLYAFGQ 423
Lambda K H
0.318 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12512387114
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40