bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1681_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
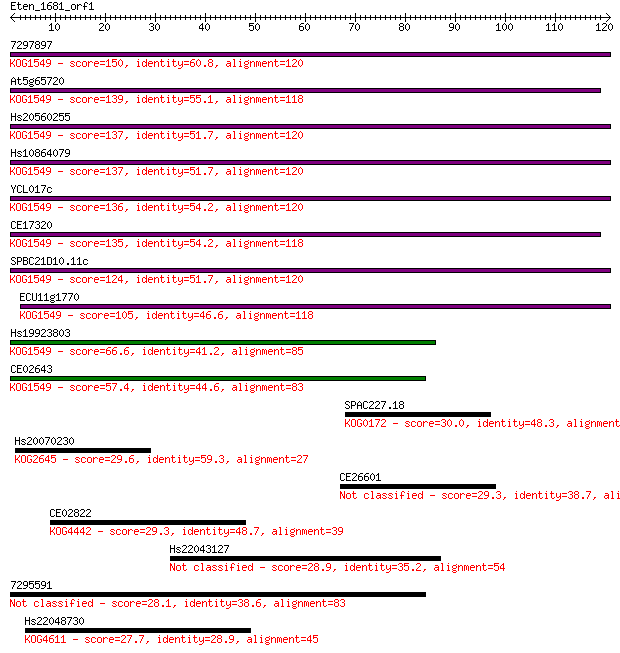
7297897 150 8e-37
At5g65720 139 2e-33
Hs20560255 137 3e-33
Hs10864079 137 3e-33
YCL017c 136 8e-33
CE17320 135 1e-32
SPBC21D10.11c 124 6e-29
ECU11g1770 105 2e-23
Hs19923803 66.6 1e-11
CE02643 57.4 7e-09
SPAC227.18 30.0 1.1
Hs20070230 29.6 1.5
CE26601 29.3 1.7
CE02822 29.3 1.7
Hs22043127 28.9 2.6
7295591 28.1 4.1
Hs22048730 27.7 5.4
> 7297897
Length=462
Score = 150 bits (378), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 73/120 (60%), Positives = 91/120 (75%), Gaps = 1/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHKIYGPKGVGAL+VR RRP+VRL PI GGGQERG+RSGT+P PL VGLG AA L+L
Sbjct 259 ISGHKIYGPKGVGALYVR-RRPRVRLEPIQSGGGQERGLRSGTVPAPLAVGLGAAAELSL 317
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
MD D++ V+ ++ LL R+ LPH+ NG Y G LN+SF++VEGESLLM++ D
Sbjct 318 REMDYDKKWVDFLSNRLLDRISSALPHVIRNGDAKATYNGCLNLSFAYVEGESLLMALKD 377
> At5g65720
Length=453
Score = 139 bits (349), Expect = 2e-33, Method: Composition-based stats.
Identities = 65/118 (55%), Positives = 93/118 (78%), Gaps = 1/118 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKGVGAL+VR RRP++RL P+++GGGQERG+RSGT T IVG G A LA+
Sbjct 250 MSAHKIYGPKGVGALYVR-RRPRIRLEPLMNGGGQERGLRSGTGATQQIVGFGAACELAM 308
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSI 118
+ M+ D + ++ + LL+ ++++L + VNGS++ RY+GNLN+SF++VEGESLLM +
Sbjct 309 KEMEYDEKWIKGLQERLLNGVREKLDGVVVNGSMDSRYVGNLNLSFAYVEGESLLMGL 366
> Hs20560255
Length=457
Score = 137 bits (346), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 62/120 (51%), Positives = 90/120 (75%), Gaps = 1/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHKIYGPKGVGA+++R RRP+VR+ + GGGQERGMRSGT+PTPL+VGLG A +A
Sbjct 254 ISGHKIYGPKGVGAIYIR-RRPRVRVEALQSGGGQERGMRSGTVPTPLVVGLGAACEVAQ 312
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ M+ D + + +++ L+ + + LP + +NG Y G +N+SF++VEGESLLM++ D
Sbjct 313 QEMEYDHKRISKLSERLIQNIMKSLPDVVMNGDPKHHYPGCINLSFAYVEGESLLMALKD 372
> Hs10864079
Length=457
Score = 137 bits (346), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 62/120 (51%), Positives = 90/120 (75%), Gaps = 1/120 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+SGHKIYGPKGVGA+++R RRP+VR+ + GGGQERGMRSGT+PTPL+VGLG A +A
Sbjct 254 ISGHKIYGPKGVGAIYIR-RRPRVRVEALQSGGGQERGMRSGTVPTPLVVGLGAACEVAQ 312
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ M+ D + + +++ L+ + + LP + +NG Y G +N+SF++VEGESLLM++ D
Sbjct 313 QEMEYDHKRISKLSERLIQNIMKSLPDVVMNGDPKHHYPGCINLSFAYVEGESLLMALKD 372
> YCL017c
Length=497
Score = 136 bits (343), Expect = 8e-33, Method: Composition-based stats.
Identities = 65/120 (54%), Positives = 91/120 (75%), Gaps = 2/120 (1%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKG+GA++VR RRP+VRL P++ GGGQERG+RSGTL PL+ G G+AA L
Sbjct 295 ISSHKIYGPKGIGAIYVR-RRPRVRLEPLLSGGGQERGLRSGTLAPPLVAGFGEAARLMK 353
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
+ D+D+ H++R++ L+ L H T+NGS + RY G +N+SF++VEGESLLM++ D
Sbjct 354 KEFDNDQAHIKRLSDKLVKGLLSA-EHTTLNGSPDHRYPGCVNVSFAYVEGESLLMALRD 412
> CE17320
Length=446
Score = 135 bits (341), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 64/118 (54%), Positives = 90/118 (76%), Gaps = 1/118 (0%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKG GAL+VR RRP+VR+ + GGGQERG+RSGT+ PL +GLG+AA +A
Sbjct 243 ISAHKIYGPKGAGALYVR-RRPRVRIEAQMSGGGQERGLRSGTVAAPLCIGLGEAAKIAD 301
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSI 118
+ M D+ HVER++++L++ + +LPHI NG Y G +N+SF++VEGESLLM++
Sbjct 302 KEMAMDKAHVERLSQMLINGISDKLPHIIRNGDARHAYPGCVNLSFAYVEGESLLMAL 359
> SPBC21D10.11c
Length=498
Score = 124 bits (310), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 62/120 (51%), Positives = 85/120 (70%), Gaps = 2/120 (1%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+S HKIYGPKG+GA +VR RRP+VRL P+I GGGQERG+RSGTL +VG G AA +
Sbjct 296 ISAHKIYGPKGIGAAYVR-RRPRVRLEPLISGGGQERGLRSGTLAPSQVVGFGTAARICK 354
Query 61 ECMDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
E M D H+ ++++ L+ L +P+ ++NG RY G +NISF++VEGESLLM + +
Sbjct 355 EEMKYDYAHISKLSQRLIDGL-LAIPYTSLNGDPKSRYPGCVNISFNYVEGESLLMGLKN 413
> ECU11g1770
Length=432
Score = 105 bits (262), Expect = 2e-23, Method: Composition-based stats.
Identities = 55/118 (46%), Positives = 82/118 (69%), Gaps = 2/118 (1%)
Query 3 GHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLALEC 62
HKIYGPKG+GAL+VR R + +I+GGGQERG+RSGT+ +PL+VG GKAA + +
Sbjct 233 AHKIYGPKGIGALYVRRRPRVRMVP-LINGGGQERGLRSGTVASPLVVGFGKAAEICSKE 291
Query 63 MDSDRRHVERMARLLLHRLQQQLPHITVNGSLNRRYLGNLNISFSFVEGESLLMSIGD 120
M D H++ +++ L + ++ + + +NGS + + G +N+SF FVEGESLLM + D
Sbjct 292 MKRDFEHIKELSKKLKNMFKKNIEGVIINGS-EKGFPGCVNVSFPFVEGESLLMHLKD 348
> Hs19923803
Length=445
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/85 (41%), Positives = 50/85 (58%), Gaps = 1/85 (1%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLAL 60
+ GHK YGP+ +GAL++R L P++ GGGQER R GT TP+I GLGKAA L
Sbjct 255 IVGHKFYGPR-IGALYIRGLGEFTPLYPMLFGGGQERNFRPGTENTPMIAGLGKAAELVT 313
Query 61 ECMDSDRRHVERMARLLLHRLQQQL 85
+ ++ H+ + L RL+ +
Sbjct 314 QNCEAYEAHMRDVRDYLEERLEAEF 338
> CE02643
Length=328
Score = 57.4 bits (137), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 49/87 (56%), Gaps = 7/87 (8%)
Query 1 VSGHKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPLIVGLGKAASLA- 59
V GHK YGP+ GAL + R+ P++ GG QE G RSGT TP+IVGLG+AA +
Sbjct 150 VVGHKFYGPRS-GALIFNPKSK--RIPPMLLGGNQESGWRSGTENTPMIVGLGEAAKVYN 206
Query 60 ---LECMDSDRRHVERMARLLLHRLQQ 83
L R++ + LLL RL+
Sbjct 207 EGFLNIESILRQNRDYFEELLLKRLRN 233
> SPAC227.18
Length=368
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 68 RHVERMARLLLHRLQQQLPHITVNGSLNR 96
RHV R RL L + Q P I V G+L R
Sbjct 176 RHVARQVRLALKKNNNQYPRILVIGALGR 204
> Hs20070230
Length=915
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 20/32 (62%), Gaps = 6/32 (18%)
Query 2 SGHKIYGPKG-----VGALFVRARRPKVRLAP 28
SGHK YGP G G+ F A+RPK ++AP
Sbjct 314 SGHK-YGPFGPEESSYGSPFTPAKRPKRKVAP 344
> CE26601
Length=1241
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 67 RRHVERMARLLLHRLQQQLPHITVNGSLNRR 97
RR+V R + RLQ LP I N ++R+
Sbjct 575 RRNVNSAMRFIKDRLQHNLPQILENSKIDRK 605
> CE02822
Length=408
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 19/43 (44%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query 9 PKGVGA-LFVRARRPKVR-LAPI--IDGGGQERGMRSGTLPTP 47
PK V L R R+PK+ API GG ++G SGT+P P
Sbjct 244 PKDVSKELKKRGRKPKMDPTAPISTTSTGGGDKGKLSGTIPVP 286
> Hs22043127
Length=276
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 5/59 (8%)
Query 33 GGQERGMRSGTLPTPLIV-----GLGKAASLALECMDSDRRHVERMARLLLHRLQQQLP 86
GGQ G++ LP P+ GL KAA+ +E S+R + ++L R LP
Sbjct 22 GGQSPGLKETQLPMPVPSDPGSPGLCKAANSRVEMYKSEREEIGGWGKVLGRRQNWHLP 80
> 7295591
Length=781
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 32/110 (29%), Positives = 44/110 (40%), Gaps = 27/110 (24%)
Query 1 VSGHKIYG-PKGVGALFVRARRPKVRLAPIIDGGGQ-----------------ERGMRSG 42
+S +KI+G P GVGAL V R +V GGG + G
Sbjct 242 LSFYKIFGYPTGVGALLVSRRGAEVFQKRRFFGGGTINYAYPHAMDYQLRETFHQRYEDG 301
Query 43 TLPTPLIVGLGKA---------ASLALECMDSDRRHVERMARLLLHRLQQ 83
TLP IVGL + + M+ RHV +A+ L +L+Q
Sbjct 302 TLPFLSIVGLLEGFRTLERLVPRTDEFSTMERISRHVFGLAKYLEDQLRQ 351
> Hs22048730
Length=995
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 23/45 (51%), Gaps = 3/45 (6%)
Query 4 HKIYGPKGVGALFVRARRPKVRLAPIIDGGGQERGMRSGTLPTPL 48
HK+ + F+ RPK ++ ++G G G+RS T+P +
Sbjct 626 HKLISQITIDVFFIDWERPKGKVLKAVEGEG---GVRSATVPVSI 667
Lambda K H
0.323 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40