bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1683_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
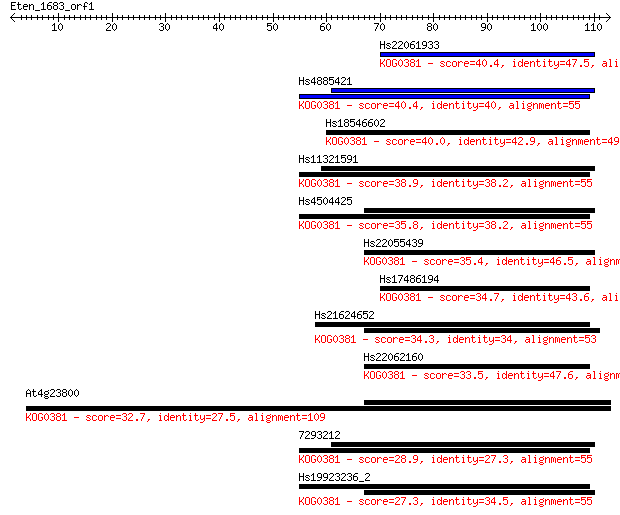
Hs22061933 40.4 7e-04
Hs4885421 40.4 8e-04
Hs18546602 40.0 0.001
Hs11321591 38.9 0.002
Hs4504425 35.8 0.018
Hs22055439 35.4 0.023
Hs17486194 34.7 0.042
Hs21624652 34.3 0.052
Hs22062160 33.5 0.100
At4g23800 32.7 0.17
7293212 28.9 2.3
Hs19923236_2 27.3 6.2
> Hs22061933
Length=411
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 27/40 (67%), Gaps = 0/40 (0%)
Query 70 EVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYK 109
+VA+ LGEMW + AA +KQ + +AK K KY +AAY+
Sbjct 328 DVAKKLGEMWNNTAADDKQPYEKRSAKLKEKYEKDIAAYR 367
> Hs4885421
Length=200
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 33/54 (61%), Gaps = 5/54 (9%)
Query 61 QPELKS-----NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYK 109
+P++KS ++ +VA+ LGEMW + +EKQ + AAK K KY +A YK
Sbjct 108 RPKIKSTNPGISIGDVAKKLGEMWNNLNDSEKQPYITKAAKLKEKYEKDVADYK 161
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 13/54 (24%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 55 QQQQQQQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++ +++ PE+ N E ++ E W++ + EK +F A +K +Y + Y
Sbjct 25 EEHKKKNPEVPVNFAEFSKKCSERWKTVSGKEKSKFDEMAKADKVRYDREMKDY 78
> Hs18546602
Length=328
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 31/49 (63%), Gaps = 2/49 (4%)
Query 60 QQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
+ P L ++ +VA+ LGEMW + AA +KQ + AAK K +Y +AAY
Sbjct 282 EHPGL--SIGDVAKKLGEMWNNTAADDKQPYEKKAAKLKEEYRKDIAAY 328
> Hs11321591
Length=209
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 34/56 (60%), Gaps = 5/56 (8%)
Query 59 QQQPELKS-----NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYK 109
+ +P++KS ++ + A+ LGEMW +A +KQ + AAK K KY +AAY+
Sbjct 108 EHRPKIKSEHPGLSIGDTAKKLGEMWSEQSAKDKQPYEQKAAKLKEKYEKDIAAYR 163
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 55 QQQQQQQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++ +++ P+ N E ++ E W++ +A EK +F A +KA+Y + Y
Sbjct 25 EEHKKKHPDSSVNFAEFSKKCSERWKTMSAKEKSKFEDMAKSDKARYDREMKNY 78
> Hs4504425
Length=215
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 67 NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYK 109
++ +VA+ LGEMW + AA +KQ + AAK K KY +AAY+
Sbjct 121 SIGDVAKKLGEMWNNTAADDKQPYEKKAAKLKEKYEKDIAAYR 163
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 55 QQQQQQQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++ +++ P+ N E ++ E W++ +A EK +F A +KA+Y + Y
Sbjct 25 EEHKKKHPDASVNFSEFSKKCSERWKTMSAKEKGKFEDMAKADKARYEREMKTY 78
> Hs22055439
Length=168
Score = 35.4 bits (80), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 67 NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYK 109
++ +VA+ LGEMW + AA +KQ + AAK K KY +AAY+
Sbjct 89 SIGDVAKKLGEMWNNTAADDKQPYEKKAAKLKEKYEKDIAAYR 131
> Hs17486194
Length=149
Score = 34.7 bits (78), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 70 EVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++ + LG MW + AA EKQ A K K KY +AAY
Sbjct 97 DIVKKLGGMWNNTAADEKQPHEKKAVKLKEKYKKDIAAY 135
> Hs21624652
Length=186
Score = 34.3 bits (77), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 58 QQQQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++QQP KE +R E WRS + EK ++ A A +KA+Y + Y
Sbjct 28 KEQQPSTYVGFKEFSRKCSEKWRSISKHEKAKYEALAKLDKARYQEEMMNY 78
Score = 30.4 bits (67), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 67 NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYKQ 110
+V +VA+ G+MW +A EK + A +AKY L Y++
Sbjct 119 SVVQVAKATGKMWSTATDLEKHPYEQRVALLRAKYFEELELYRK 162
> Hs22062160
Length=187
Score = 33.5 bits (75), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 67 NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++ +VA+ LGEMW + AA +KQ AAK K KY +AAY
Sbjct 97 SIGDVAKKLGEMWNNTAADDKQPGEKKAAKLKEKYEKDIAAY 138
> At4g23800
Length=456
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 67 NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYKQHK 112
+V EVA++ GE W++ + +K + A K K Y A+ YK+ K
Sbjct 279 SVVEVAKITGEEWKNLSDKKKAPYEKVAKKNKETYLQAMEEYKRTK 324
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 51/113 (45%), Gaps = 15/113 (13%)
Query 4 PRLGFAGGLEYDEQQEQQPARRGQFRCCSS----SNESVIWQQQEQQQQQQRAPEQQQQQ 59
P + FA G Q EQ+ A + + + C S+ V+W + + + ++ PE
Sbjct 108 PNMTFACGQSSLTQAEQEKANKKKKKDCPETKRPSSSYVLWCKDQWTEVKKENPE----- 162
Query 60 QQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYKQHK 112
++ KE + +LG W+S +A +K+ + EK Y +A K+ K
Sbjct 163 ------ADFKETSNILGAKWKSLSAEDKKPYEERYQVEKEAYLQVIAKEKREK 209
> 7293212
Length=328
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 61 QPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYK 109
PE V ++A+ LG W KQ++ + A ++KA+Y + YK
Sbjct 228 NPEF--GVGDIAKELGRKWSDVDPEVKQKYESMAERDKARYEREMTEYK 274
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 55 QQQQQQQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++ +++ P+ E +R E W++ EK++F A K+K +Y A + Y
Sbjct 130 EEHKKKHPDETVIFAEFSRKCAERWKTMVDKEKKRFHEMAEKDKQRYEAEMQNY 183
> Hs19923236_2
Length=181
Score = 27.3 bits (59), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 67 NVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAYK 109
++ +V + L MW + AAA+KQ + AAK K KY +AAY+
Sbjct 97 SIDDVVKKLAGMWNNTAAADKQFYEKKAAKLKEKYKKDIAAYR 139
Score = 27.3 bits (59), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 13/54 (24%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 55 QQQQQQQPELKSNVKEVARVLGEMWRSAAAAEKQQFAAAAAKEKAKYAAALAAY 108
++ +++ P+ E + E W++ A EK +F A +KA Y + Y
Sbjct 1 EEHKKKNPDASVKFSEFLKKCSETWKTIFAKEKGKFEDMAKADKAHYEREMKTY 54
Lambda K H
0.310 0.119 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1188972946
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40