bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1685_orf1
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
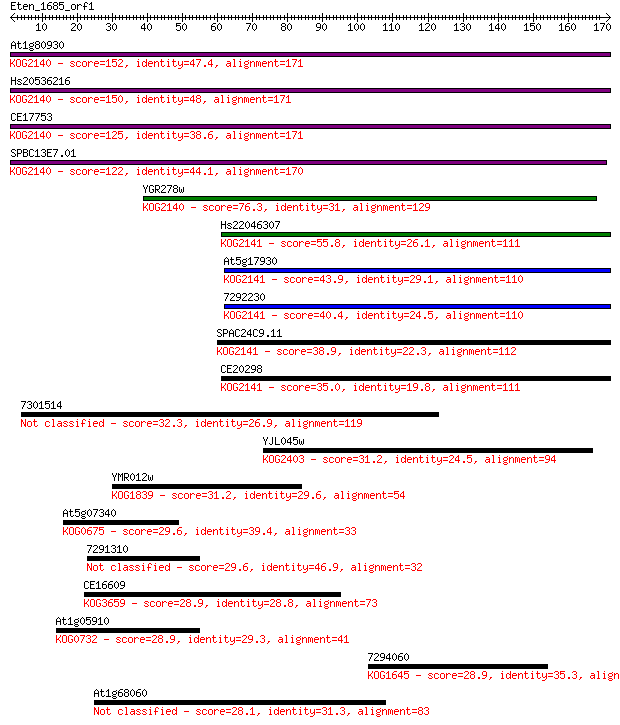
At1g80930 152 4e-37
Hs20536216 150 1e-36
CE17753 125 3e-29
SPBC13E7.01 122 3e-28
YGR278w 76.3 3e-14
Hs22046307 55.8 4e-08
At5g17930 43.9 2e-04
7292230 40.4 0.002
SPAC24C9.11 38.9 0.005
CE20298 35.0 0.075
7301514 32.3 0.48
YJL045w 31.2 1.0
YMR012w 31.2 1.2
At5g07340 29.6 2.7
7291310 29.6 3.3
CE16609 28.9 4.5
At1g05910 28.9 4.8
7294060 28.9 5.5
At1g68060 28.1 8.5
> At1g80930
Length=900
Score = 152 bits (383), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 81/172 (47%), Positives = 118/172 (68%), Gaps = 4/172 (2%)
Query 1 DEKKWAALSREILG-EESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVV 59
+EKK+ AL +E+LG EES D D S SE E+E++E ++Q ++D T+ +V
Sbjct 592 NEKKYEALKKELLGDEESEDEDGSDASSEDNDEEEDESDEED---EEQMRIRDETETNLV 648
Query 60 CLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAER 119
LR+TIYL IMSS++ EE HK+LK+ ++ EME+ +ML++CCS ERT+ R++ L +R
Sbjct 649 NLRRTIYLTIMSSVDFEEAGHKLLKIKLEPGQEMELCIMLLECCSQERTYLRYYGLLGQR 708
Query 120 LAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ + E F++ F +QY++ HRLET KLRN AKFFAHLL +DA+PW VL
Sbjct 709 FCMINKIHQENFEKCFVQQYSMIHRLETNKLRNVAKFFAHLLGTDALPWHVL 760
> Hs20536216
Length=908
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 82/172 (47%), Positives = 110/172 (63%), Gaps = 1/172 (0%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTV-VKDLTDQEVV 59
+E+K+ A+ +EIL E TD++ + +E EEE EE E + Q V + D T+ +V
Sbjct 394 NEEKYKAIKKEILDEGDTDSNTDQDAGSSEEDEEEEEEEGEEDEEGQKVTIHDKTEINLV 453
Query 60 CLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAER 119
R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT+++FF L A R
Sbjct 454 SFRRTIYLAIQSSLDFEECAHKLLKMEFPESQTKELCNMILDCCAQQRTYEKFFGLLAGR 513
Query 120 LAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
LK Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSVL
Sbjct 514 FCMLKKEYMESFEGIFKEQYDTIHRLETNKLRNVAKMFAHLLYTDSLPWSVL 565
> CE17753
Length=897
Score = 125 bits (315), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 66/171 (38%), Positives = 105/171 (61%), Gaps = 4/171 (2%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVVC 60
+E+ + + +EI+G +AD S ED E EEE + E KK T + D TDQ +
Sbjct 430 NEEVYEEIRKEIIG----NADISDEDGGDELDDEEEGSDVEEAPKKTTEIIDNTDQNLTA 485
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
R+ +YL + SSL+ +E HK+LKM I + ++ E+ ML+DCC+ +RT++RF+ + ER
Sbjct 486 FRREVYLTMQSSLDYQEAAHKLLKMKIPDSMQNELCAMLVDCCAQQRTYERFYGMLIERF 545
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+L+ Y + F++ + Y+ HR++ KLRN A+ AHLL +DAI W +L
Sbjct 546 CRLRLEYQQYFEKLCQDTYSTIHRIDITKLRNLARLIAHLLSTDAIDWKIL 596
> SPBC13E7.01
Length=628
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 75/170 (44%), Positives = 108/170 (63%), Gaps = 3/170 (1%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVVC 60
+EKK+ A+ EILGEE D + E+ E+S EE E + +K V+ D T+ +V
Sbjct 355 NEKKYDAIKHEILGEEDDDENEEDEEDSEETSESEEDESVND--EKPQVI-DQTNASLVN 411
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
LRK+IYL IMSS++ EEC HK+LK+++ E E+E+ M+I+C S ERT+ +F+ L ER
Sbjct 412 LRKSIYLTIMSSVDFEECCHKLLKIDLPEGQEIELCNMVIECNSQERTYAKFYGLIGERF 471
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
KL + +++ FK Y HR ET +LRN A FFA+LL +D+I W V
Sbjct 472 CKLSRTWRSTYEQCFKNYYETIHRYETNRLRNIALFFANLLSTDSIGWEV 521
> YGR278w
Length=577
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 69/130 (53%), Gaps = 1/130 (0%)
Query 39 ETAEPGKKQTVVKDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVM 98
+T E Q + D+T V +K IYL + SSL+ +E HK+LK+ I +L+ V +
Sbjct 269 DTNEGSNSQLQIYDMTSTNDVEFKKKIYLVLKSSLSGDEAAHKLLKLKIANNLKKSVVDI 328
Query 99 LIDCCSMERTFQRFFALQAERLAKLKTAYCECFQEAFKRQYAL-AHRLETAKLRNTAKFF 157
+I E TF +F+++ +ER+ ++ + E F++ Y ET +LR KF+
Sbjct 329 IIKSSLQESTFSKFYSILSERMITFHRSWQTAYNETFEQNYTQDIEDYETDQLRILGKFW 388
Query 158 AHLLMSDAIP 167
HL+ + +P
Sbjct 389 GHLISYEFLP 398
> Hs22046307
Length=356
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 57/111 (51%), Gaps = 0/111 (0%)
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
+R+ I+ IM+S + + K+LK+ +++ E E+ +L+DCC E+T+ F+A A +
Sbjct 151 IRRNIFCTIMTSEDFLDAFEKLLKLGLKDQQEREIIHVLMDCCLQEKTYNPFYAFLASKF 210
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ + + FQ + ++ L N AHLL + ++ S+L
Sbjct 211 CEYERRFQMTFQFSIWDKFRDLENLPATNFSNLVHLVAHLLKTKSLSLSIL 261
> At5g17930
Length=707
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 60/119 (50%), Gaps = 18/119 (15%)
Query 62 RKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL- 120
RK I+ IMSS + + K+L++++ + E+ +L++CC E+ F +F+ + A +L
Sbjct 510 RKAIFCVIMSSEDYIDAFEKLLRLDLPGKQDREIMRVLVECCLQEKAFNKFYTVLASKLC 569
Query 121 -----AKLKTAYCECFQEAFKRQYALAHRLETAKLRNT---AKFFAHLLMSDAIPWSVL 171
K Y C + FK LE+ L+ + AKF A ++++ + +VL
Sbjct 570 EHDKNHKFTLQY--CIWDHFK-------ELESMSLQRSMHLAKFVAEIIVTFNLSLAVL 619
> 7292230
Length=854
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/110 (24%), Positives = 54/110 (49%), Gaps = 2/110 (1%)
Query 62 RKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERLA 121
R+ I+ IMS+ + + KIL +++++ + VA ++I C E+ ++A A +
Sbjct 652 RRNIFCIIMSAADYVDAFEKILHLSLKD--QRAVAYVIIHCALNEKRANPYYAHLALKFC 709
Query 122 KLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ Y FQ A + +L ++RN A F ++++ + SVL
Sbjct 710 QFNRKYQLAFQFASWDRINDIEKLSKPQIRNLASFLQQVILAAGLQLSVL 759
> SPAC24C9.11
Length=775
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 25/120 (20%), Positives = 58/120 (48%), Gaps = 8/120 (6%)
Query 60 CLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAER 119
LR +I++ ++ S + + ++LK++++ + E+A +++ C E+ + F+ L A +
Sbjct 565 TLRTSIFVALVGSEDYIDAWERVLKLHLKRNQLPEIAYVILHCVGNEKLYNPFYGLVALK 624
Query 120 LAKLKTAYCECFQ--------EAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
L+ + FQ E + + ++ N AK +A L++ A P ++L
Sbjct 625 CCTLQHNLKKSFQFSLWDFFNELQPDDDSEEREISMRRIVNLAKLYASLVIEAAQPLTIL 684
> CE20298
Length=819
Score = 35.0 bits (79), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 22/111 (19%), Positives = 55/111 (49%), Gaps = 0/111 (0%)
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
+R+ I+ + S+ + ++ ++LK+ ++ + E E+ +LI E+T+ F+A +R
Sbjct 628 VRRHIFCSVASADDEDDAFERLLKLQLKGEKERELVHVLIAMMMKEKTYNAFYATLLQRF 687
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ + Q A + +L+ + + A+ HL+ ++ + +VL
Sbjct 688 CEFNKRFVITLQFALWDRLRECDQLKPFQRGSLAQLLQHLISNEVMSITVL 738
> 7301514
Length=167
Score = 32.3 bits (72), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 32/130 (24%), Positives = 59/130 (45%), Gaps = 18/130 (13%)
Query 4 KWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVVCLRK 63
+WA+L+R++ G S D DASS+D+ A+ ++ + G KD T + LR
Sbjct 21 RWASLARQLSGSSSQD-DASSKDAAAQEAASGCPPNSTTTGTDTPKAKDKTTKGRKRLRN 79
Query 64 T-----IYLCIMSS------LNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRF 112
C MS L+ + + K+L N +E E+ ++ + + + F
Sbjct 80 IEIPPEPTTCCMSGCANCVWLDYAQTLAKLLGDNDEEAREIVLSKI------TDPNLKMF 133
Query 113 FALQAERLAK 122
+L+ ++AK
Sbjct 134 LSLELRQMAK 143
> YJL045w
Length=634
Score = 31.2 bits (69), Expect = 1.0, Method: Composition-based stats.
Identities = 23/94 (24%), Positives = 39/94 (41%), Gaps = 1/94 (1%)
Query 73 LNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERLAKLKTAYCECFQ 132
L ++ +H + + + +E+E M R +QR F Q++ K AY C
Sbjct 118 LGDQDAIHYMTREAPKSVIELEHYGMPFSRTEDGRIYQRAFGGQSKDFGKGGQAYRTC-A 176
Query 133 EAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAI 166
A + +A+ H L L+N FF D +
Sbjct 177 VADRTGHAMLHTLYGQALKNNTHFFIEYFAMDLL 210
> YMR012w
Length=1277
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 32/54 (59%), Gaps = 2/54 (3%)
Query 30 ESSSEEEAEETAEPGKKQTVVKDLTDQEVVCLRKTIYLCIMSSLNAEECVHKIL 83
E S +EE +E ++P +K+ KD+TD+E + + ++ S N+ ++K+L
Sbjct 155 ERSKQEEKDEKSDPEEKKNTFKDVTDEEKLKFNEMVHEVFSSFKNSS--INKLL 206
> At5g07340
Length=532
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 16 ESTDADASSEDSEAESSSEEEAEETAEPGKKQT 48
E + ++E S +E+ +EE+AE A P K+QT
Sbjct 496 EKKKPETAAETSTSEAKTEEKAEAVAAPRKRQT 528
> 7291310
Length=1566
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 23 SSEDSEAESSSEEEAEETAEPGKKQTVVKDLT 54
S E++E ES S AEETA ++Q K+LT
Sbjct 83 SVENAENESDSGGNAEETAAADRRQATKKELT 114
> CE16609
Length=637
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 38/76 (50%), Gaps = 4/76 (5%)
Query 22 ASSEDSEAESSSEEEAEETAEPGKKQTVVKD---LTDQEVVCLRKTIYLCIMSSLNAEEC 78
ASS D + + ++EET +TV D T +E++ ++KT +S++ +
Sbjct 2 ASSSDFKQLKQTRTKSEETTRSASNETVNDDNRSTTTEEIIPIKKTSNATAISNIGFKSV 61
Query 79 VHKILKMNIQEDLEME 94
+ L+ NI D +ME
Sbjct 62 TNLALE-NIGLDTKME 76
> At1g05910
Length=1069
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 14 GEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLT 54
G+E DA++ S A S ++ + E+ +EP K+ +D++
Sbjct 947 GQEMPSPDAANPQSAAPSPTDGDREDQSEPPSKEASAEDMS 987
> 7294060
Length=608
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 103 CSMERTFQRFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNT 153
C + TF+ L A+R+ L T E + +R + L L TAKL NT
Sbjct 179 CKTKATFRDIRHLYAKRIQMLDTDIREQLEAERRRTHTLTTELATAKLANT 229
> At1g68060
Length=622
Score = 28.1 bits (61), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 44/87 (50%), Gaps = 8/87 (9%)
Query 25 EDSEAESSSEEEAEETAE--PGKKQTVVKDLTDQEVVCLRKTIYLCIMSSLNAEECVHKI 82
EDSE + SE A T + PG V+ DL +EVV LRK+ + S + ++ + +
Sbjct 494 EDSEEKPPSELPAPATEDNVPG----VLYDLLQKEVVALRKSSHEKDQSLKDKDDAIEML 549
Query 83 LKM--NIQEDLEMEVAVMLIDCCSMER 107
K + + +E+E M + +ME+
Sbjct 550 AKKVETLTKAMEVEAKKMRREVAAMEK 576
Lambda K H
0.317 0.127 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2526689620
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40