bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1686_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
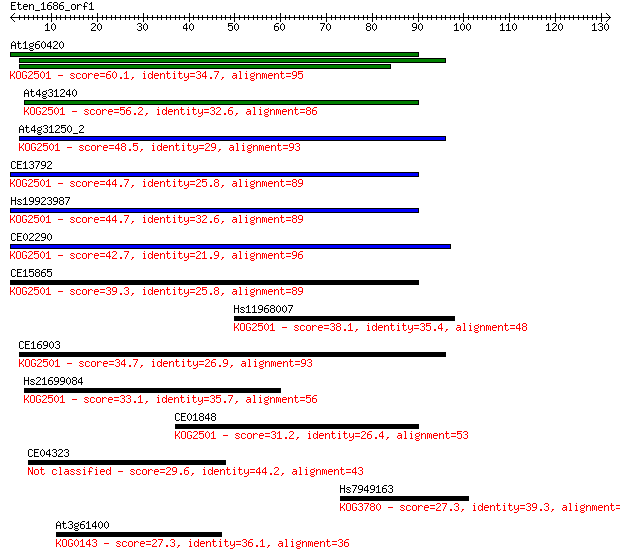
At1g60420 60.1 1e-09
At4g31240 56.2 2e-08
At4g31250_2 48.5 3e-06
CE13792 44.7 4e-05
Hs19923987 44.7 5e-05
CE02290 42.7 2e-04
CE15865 39.3 0.002
Hs11968007 38.1 0.004
CE16903 34.7 0.053
Hs21699084 33.1 0.15
CE01848 31.2 0.52
CE04323 29.6 1.7
Hs7949163 27.3 7.5
At3g61400 27.3 8.7
> At1g60420
Length=578
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 7/91 (7%)
Query 1 PHCRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTMPWLSLPFGD- 59
P CR F+ +L +VY Q+K+ + FE I+I DR F + MPWL+LPFGD
Sbjct 376 PPCRAFTPKLVEVYKQIKE-----RNEAFELIFISSDRDQESFDEYYSQMPWLALPFGDP 430
Query 60 -YCALVRRFGIRRLPSIILITPDDHILTSDA 89
+L + F + +P + + P +T +A
Sbjct 431 RKASLAKTFKVGGIPMLAALGPTGQTVTKEA 461
Score = 47.8 bits (112), Expect = 6e-06, Method: Composition-based stats.
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 7/95 (7%)
Query 3 CRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTMPWLSLPFGDYCA 62
C + + +L + Y++LK+ + + FE + I + F T PWL+LPF D
Sbjct 218 CTELTPKLVEFYTKLKENKED-----FEIVLISLEDDEESFNQDFKTKPWLALPFNDKSG 272
Query 63 --LVRRFGIRRLPSIILITPDDHILTSDAVPIFDD 95
L R F + LP+++++ PD S+ DD
Sbjct 273 SKLARHFMLSTLPTLVILGPDGKTRHSNVAEAIDD 307
Score = 45.4 bits (106), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/84 (29%), Positives = 44/84 (52%), Gaps = 11/84 (13%)
Query 3 CRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTMPWLSLPFGDYCA 62
C+ F+ +L +VY++L G FE +++ D F + MPWL++PF D
Sbjct 58 CQRFTPQLVEVYNELSSKVG------FEIVFVSGDEDEESFGDYFRKMPWLAVPFTDSET 111
Query 63 ---LVRRFGIRRLPSIILITPDDH 83
L F +R +P+++++ DDH
Sbjct 112 RDRLDELFKVRGIPNLVMV--DDH 133
> At4g31240
Length=204
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 48/88 (54%), Gaps = 6/88 (6%)
Query 4 RDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTMPWLSLPFGDYCA- 62
R F+++L VY++L FE I I DR EF MPWL++P+ D
Sbjct 31 RSFTSQLVDVYNEL----ATTDKGSFEVILISTDRDSREFNINMTNMPWLAIPYEDRTRQ 86
Query 63 -LVRRFGIRRLPSIILITPDDHILTSDA 89
L R F ++ +P++++I P++ +T++A
Sbjct 87 DLCRIFNVKLIPALVIIGPEEKTVTTNA 114
> At4g31250_2
Length=281
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 49/95 (51%), Gaps = 7/95 (7%)
Query 3 CRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTMPWLSLPFGDYC- 61
C+DF+ L ++Y L+ +GE+ E I++ D F MPWL++PF
Sbjct 138 CKDFTPELIKLYENLQN-RGEE----LEIIFVSFDHDMTSFYEHFWCMPWLAVPFNLSLL 192
Query 62 -ALVRRFGIRRLPSIILITPDDHILTSDAVPIFDD 95
L ++GI R+PS++ + D+ + D + + +D
Sbjct 193 NKLRDKYGISRIPSLVPLYSDEISVAEDVIGLIED 227
> CE13792
Length=155
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 44/93 (47%), Gaps = 12/93 (12%)
Query 1 PHCRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFA--CTMPWLSLPFG 58
P CR F+ L Y ++ + FE +++ DRS ++ + C W +P G
Sbjct 52 PPCRGFTPILKDFYEEVNE--------EFEIVFVSSDRSESDLKMYMKECHGDWYHIPHG 103
Query 59 DYCA--LVRRFGIRRLPSIILITPDDHILTSDA 89
+ L ++G+ +P++I++ PD +T D
Sbjct 104 NGAKQKLSTKYGVSGIPALIIVKPDGTEITRDG 136
> Hs19923987
Length=212
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 44/93 (47%), Gaps = 11/93 (11%)
Query 1 PHCRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTMP--WLSLPFG 58
P +DF RL + L+ Q +Y+ D + + F MP WL LPF
Sbjct 52 PILKDFFVRLTDEFYVLRAAQ-------LALVYVSQDSTEEQQDLFLKDMPKKWLFLPFE 104
Query 59 DYCA--LVRRFGIRRLPSIILITPDDHILTSDA 89
D L R+F + RLP+++++ PD +LT D
Sbjct 105 DDLRRDLGRQFSVERLPAVVVLKPDGDVLTRDG 137
> CE02290
Length=149
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/100 (21%), Positives = 50/100 (50%), Gaps = 9/100 (9%)
Query 1 PHCRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAF--ACTMPWLSLPFG 58
P CR F+ +L + + +++ ++ P FE +++ DR + + W ++PFG
Sbjct 39 PPCRQFTPKLTRFFDEIR-----KKHPEFEVVFVSRDREDGDLREYFLEHMGAWTAIPFG 93
Query 59 D--YCALVRRFGIRRLPSIILITPDDHILTSDAVPIFDDR 96
L+ ++ ++ +PS+ ++ P+ ++ DA D+
Sbjct 94 TPRIQELLEQYEVKTIPSMRIVKPNGDVVVQDARTEIQDK 133
> CE15865
Length=777
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 23/91 (25%), Positives = 40/91 (43%), Gaps = 10/91 (10%)
Query 1 PHCRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTM--PWLSLPFG 58
P RDF+ L Q YSQ++ FE +++ D + E + W LP
Sbjct 672 PPSRDFTPVLAQFYSQVED--------NFEILFVSSDNNTQEMNFYLQNFHGDWFHLPLN 723
Query 59 DYCALVRRFGIRRLPSIILITPDDHILTSDA 89
++ R +P++I++ PD ++T D
Sbjct 724 LCNSMKHRNTKNHIPALIIMKPDGTVITDDG 754
> Hs11968007
Length=186
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 31/52 (59%), Gaps = 4/52 (7%)
Query 50 MPWLSLPFGDYC---ALVRRFGIRRLPSIILITPDDHILTSDA-VPIFDDRD 97
MPWL++P+ D L R +GI+ +P++I++ P ++T V + +D D
Sbjct 1 MPWLAVPYTDEARRSRLNRLYGIQGIPTLIMLDPQGEVITRQGRVEVLNDED 52
> CE16903
Length=151
Score = 34.7 bits (78), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 25/107 (23%), Positives = 45/107 (42%), Gaps = 19/107 (17%)
Query 3 CRDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAF-ACTMPWLSLPFGD-- 59
CR F+ +L + Y LK E E + + DR + + W+++PFGD
Sbjct 42 CRQFTPKLKRFYEALKAAGKE-----IEVVLVSRDREAEDLLEYLGHGGDWVAIPFGDER 96
Query 60 YCALVRRFGIRRLPSIILITPDDHIL-----------TSDAVPIFDD 95
++++ + +P+ LI +L DAV +FD+
Sbjct 97 IQEYLKKYEVPTIPAFKLINNAGELLHDARADVTERGKDDAVALFDE 143
> Hs21699084
Length=135
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query 4 RDFSARLHQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAFACTM--PWLSLPFGD 59
RDF+ L Y+ L + ++P PFE +++ D S E F + WL+LPF D
Sbjct 42 RDFTPLLCDFYTAL--VAEARRPAPFEVVFVSADGSCQEMLDFMRELHGAWLALPFHD 97
> CE01848
Length=140
Score = 31.2 bits (69), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 14/56 (25%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Query 37 DRSPAEFAAFACTMP-WLSLPFGDYC--ALVRRFGIRRLPSIILITPDDHILTSDA 89
DRS ++ + W +P+G+ L ++G+ +P++I++ PD +T D
Sbjct 66 DRSESDLKMYMSEHGDWYHIPYGNDAIKELSTKYGVSGIPALIIVKPDGTEVTKDG 121
> CE04323
Length=283
Score = 29.6 bits (65), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 23/45 (51%), Gaps = 5/45 (11%)
Query 5 DFSARLHQVYSQLKQ--LQGEQQPPPFETIYIPCDRSPAEFAAFA 47
DFS LH SQL G +Q P FET++ P PA +A A
Sbjct 10 DFSQPLHYALSQLNADLYAGIEQEPHFETVHFP---KPASLSAMA 51
> Hs7949163
Length=391
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 73 PSIILITPDDHILTSDAVPIFDDRDAAQ 100
P + + P+DH L S P+ DD D +Q
Sbjct 332 PCYMDVIPEDHRLESPTTPLLDDMDGSQ 359
> At3g61400
Length=370
Score = 27.3 bits (59), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 6/36 (16%)
Query 11 HQVYSQLKQLQGEQQPPPFETIYIPCDRSPAEFAAF 46
H+VY +K+L EQ PP + D + +EFA+
Sbjct 325 HRVYGPIKELLSEQNPPKYR------DTTISEFASM 354
Lambda K H
0.326 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40