bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1707_orf3
Length=287
Score E
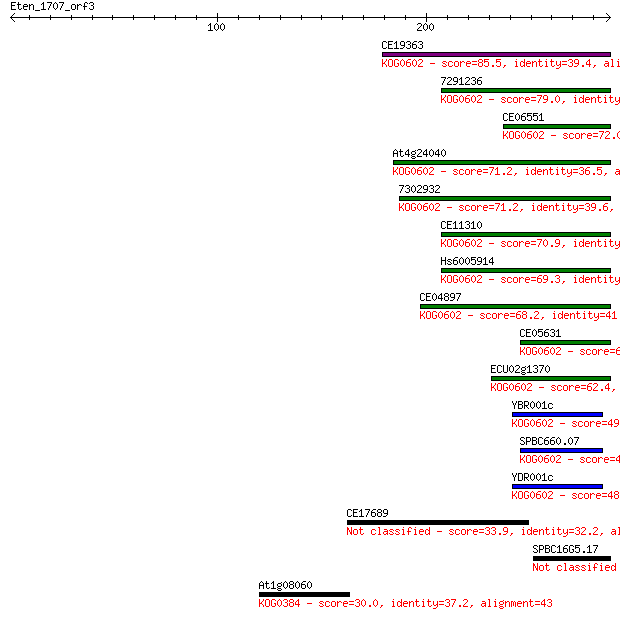
Sequences producing significant alignments: (Bits) Value
CE19363 85.5 1e-16
7291236 79.0 1e-14
CE06551 72.0 1e-12
At4g24040 71.2 2e-12
7302932 71.2 2e-12
CE11310 70.9 3e-12
Hs6005914 69.3 9e-12
CE04897 68.2 2e-11
CE05631 63.2 7e-10
ECU02g1370 62.4 1e-09
YBR001c 49.7 7e-06
SPBC660.07 48.1 2e-05
YDR001c 48.1 2e-05
CE17689 33.9 0.41
SPBC16G5.17 31.6 1.8
At1g08060 30.0 5.1
> CE19363
Length=684
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 43/109 (39%), Positives = 65/109 (59%), Gaps = 10/109 (9%)
Query 179 LPGEKGDTATVKNSEIMGENEFARRAVLLWAKLGRKTRRTVKTGSPIGSEASDNKIVSSS 238
LP + T + N + FA+R +W +L R+ + VK ++ S
Sbjct 168 LPDWRPITEQLANIKDASYQAFAQRLHFIWIQLCRQIKPEVK----------NDPSRFSL 217
Query 239 VHVPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
++VP+ FI+PGGRFRE YYWD+YWI++GL+ S + STAR +ILNFA ++
Sbjct 218 IYVPYQFILPGGRFREFYYWDAYWILKGLIASELYSTARMMILNFAHII 266
> 7291236
Length=596
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 48/81 (59%), Gaps = 10/81 (12%)
Query 207 LWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRG 266
+W LGRK + E S N S + VP IVPGGRF E YYWDSYWI+RG
Sbjct 159 IWKDLGRKMK----------DEVSKNPEYYSIIPVPNPVIVPGGRFIEFYYWDSYWIIRG 208
Query 267 LLHSGMISTARGIILNFADLV 287
LL+S M TARG+I NF +V
Sbjct 209 LLYSQMFDTARGMIENFFSIV 229
> CE06551
Length=588
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 30/51 (58%), Positives = 39/51 (76%), Gaps = 0/51 (0%)
Query 237 SSVHVPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
S ++VP FIVPGGRFRE YYWD+YWI++GL+ S M +T R +I N A +V
Sbjct 142 SLLYVPNSFIVPGGRFREFYYWDAYWIIKGLIASDMYNTTRSMIRNLASMV 192
> At4g24040
Length=626
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 38/104 (36%), Positives = 60/104 (57%), Gaps = 13/104 (12%)
Query 184 GDTATVKNSEIMGENEFARRAVLLWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPW 243
G + V+N E+ E+AR LW L + +V+ E++D + + +P
Sbjct 170 GFLSNVENEEV---REWAREVHGLWRNLSCRVSDSVR-------ESADRHTL---LPLPE 216
Query 244 GFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
I+PG RFRE+YYWDSYW+++GL+ S M +TA+G++ N LV
Sbjct 217 PVIIPGSRFREVYYWDSYWVIKGLMTSQMFTTAKGLVTNLMSLV 260
> 7302932
Length=332
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 54/101 (53%), Gaps = 13/101 (12%)
Query 187 ATVKNSEIMGENEFARRAVLLWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFI 246
A + + EI +F LW +LGR+ I E +N S ++VP FI
Sbjct 71 ARISDPEI---KQFGSDVNGLWKELGRR----------IKDEVKENPDQYSIIYVPNPFI 117
Query 247 VPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
VP RE YW+S+WI+RGLL GM TARG+I N+ +LV
Sbjct 118 VPSSNCREYRYWESFWIIRGLLQCGMHQTARGMIDNYLELV 158
> CE11310
Length=570
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 48/81 (59%), Gaps = 10/81 (12%)
Query 207 LWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRG 266
+W L RK R VK D+ S ++VP FI+PGGRF E YYWD++WI++G
Sbjct 111 IWKDLCRKVRDDVK-------HRQDH---YSLLYVPHPFIIPGGRFLEFYYWDTFWILKG 160
Query 267 LLHSGMISTARGIILNFADLV 287
LL S M TARG+I N +V
Sbjct 161 LLFSEMYETARGVIKNLGYMV 181
> Hs6005914
Length=583
Score = 69.3 bits (168), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 46/81 (56%), Gaps = 10/81 (12%)
Query 207 LWAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRG 266
LW KLG+K + E + S ++ FIVPGGRF E YYWDSYW++ G
Sbjct 134 LWKKLGKKMK----------PEVLSHPERFSLIYSEHPFIVPGGRFVEFYYWDSYWVMEG 183
Query 267 LLHSGMISTARGIILNFADLV 287
LL S M T +G++ NF DLV
Sbjct 184 LLLSEMAETVKGMLQNFLDLV 204
> CE04897
Length=585
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/94 (40%), Positives = 54/94 (57%), Gaps = 13/94 (13%)
Query 197 ENEFARRAVLL---WAKLGRKTRRTVKTGSPIGSEASDNKIVSSSVHVPWGFIVPGGRFR 253
+ ++ R A L W L RK + V+ N S + VP F+VPGGRFR
Sbjct 93 DEDYRRFAAALHAKWPTLYRKISKKVRV----------NPEKYSIIPVPNPFVVPGGRFR 142
Query 254 EMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
EMYYWDS++ ++GL+ SGM++T +G+I N LV
Sbjct 143 EMYYWDSFFTIKGLIASGMLTTVKGMIENMIYLV 176
> CE05631
Length=638
Score = 63.2 bits (152), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 26/43 (60%), Positives = 34/43 (79%), Gaps = 0/43 (0%)
Query 245 FIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
FIVPGGRF +YWD++WI++GLL S M T +GII NF++LV
Sbjct 155 FIVPGGRFDVYFYWDTFWIIKGLLVSRMFETTKGIINNFSNLV 197
> ECU02g1370
Length=659
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 37/57 (64%), Gaps = 0/57 (0%)
Query 231 DNKIVSSSVHVPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
D S+ + V F++PG RF+E YYWD+YWI+ GL+ SGM A G++ NF L+
Sbjct 170 DKGYYSTMIKVKNPFVIPGDRFKECYYWDTYWIIEGLVKSGMHKIAVGMVENFILLI 226
> YBR001c
Length=780
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 241 VPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNF 283
V + + VPGGRF E+Y WDSY + GL+ S + ARG++ +F
Sbjct 321 VGYPYAVPGGRFNELYGWDSYLMALGLIESNKVDVARGMVEHF 363
> SPBC660.07
Length=735
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 27/39 (69%), Gaps = 0/39 (0%)
Query 245 FIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNF 283
F+VPGGRF E+Y WDSY+ GLL + A+G++ NF
Sbjct 277 FVVPGGRFNELYGWDSYFESLGLLVDDRVDLAKGMVENF 315
> YDR001c
Length=751
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 241 VPWGFIVPGGRFREMYYWDSYWIVRGLLHSGMISTARGIILNF 283
+ + + VPGGRF E+Y WDSY + GLL + ARG++ +F
Sbjct 292 IGYPYAVPGGRFNELYGWDSYMMALGLLEANKTDVARGMVEHF 334
> CE17689
Length=813
Score = 33.9 bits (76), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 43/90 (47%), Gaps = 12/90 (13%)
Query 162 PDISVCPTYTYGLPSPVLPGEKGDTATVKNSEIM-GENEFARRAVLLWA--KLGRKTRRT 218
P+ +V P YT+G P + G+ +T+T N I + F LLW + ++TR T
Sbjct 392 PNFNV-PEYTHGNPHIYVGGDMLETSTAANDPIFWMHHSFVD---LLWEMYRQSKQTRAT 447
Query 219 VKTGSPIGSEASDNKIVSSSVHVPWGFIVP 248
+T P +DN+ SS H F+ P
Sbjct 448 RETAYP-----ADNRQCSSEHHFRAAFMRP 472
> SPBC16G5.17
Length=560
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 251 RFREMYYWDSYWIVRGLLHSGMISTARGIILNFADLV 287
FR ++ +W VR L+ G I+ +R ++ N+ D+V
Sbjct 397 HFRYLFLKTVFWTVRKNLYQGFITVSRTLVPNYPDIV 433
> At1g08060
Length=1697
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 120 LAESLPSWLTDTCPGVPEATRLTPSLRQKTPYNLTPSSPRVLP 162
LA S+P+ + PG+ ++R TPS + TP + T S R+ P
Sbjct 19 LAASIPASVEQETPGLRRSSRGTPSTKVITPASATRKSERLAP 61
Lambda K H
0.320 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6375248280
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40