bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1743_orf2
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
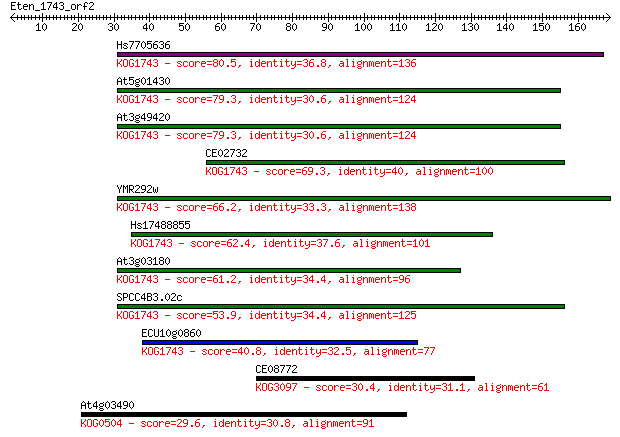
Hs7705636 80.5 1e-15
At5g01430 79.3 3e-15
At3g49420 79.3 3e-15
CE02732 69.3 3e-12
YMR292w 66.2 3e-11
Hs17488855 62.4 5e-10
At3g03180 61.2 9e-10
SPCC4B3.02c 53.9 2e-07
ECU10g0860 40.8 0.001
CE08772 30.4 1.5
At4g03490 29.6 2.8
> Hs7705636
Length=138
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 50/136 (36%), Positives = 84/136 (61%), Gaps = 1/136 (0%)
Query 31 LSDGQRIGVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKP 90
L+D Q+IG+GL G+ F GM+ F D+ALL +GN+ F++G+ V+G +T +FF +
Sbjct 4 LTDTQKIGMGLTGFGVFFLFFGMILFFDKALLAIGNVLFVAGLAFVIGLERTFRFF-FQK 62
Query 91 HKWKASLLYILGVLLIALGYSLIGLPLQLYGLLRLFASFLPQVLGAVRLSPIGCWILQLP 150
HK KA+ ++ GV ++ +G+ LIG+ ++YG LF F P V+G +R P+ +L LP
Sbjct 63 HKMKATGFFLGGVFVVLIGWPLIGMIFEIYGFFLLFRGFFPVVVGFIRRVPVLGSLLNLP 122
Query 151 GLKQCTKWVLDGDKQL 166
G++ V + + +
Sbjct 123 GIRSFVDKVGESNNMV 138
> At5g01430
Length=140
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 76/124 (61%), Gaps = 1/124 (0%)
Query 31 LSDGQRIGVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKP 90
++D ++IG+GL G+ F+G++ D+ LL +GN+ F+SG+ L +G + T++FF +K
Sbjct 6 MNDRKKIGLGLTGFGVFFSFLGIVFVFDKGLLAMGNILFISGVSLTIGFKSTMQFF-MKR 64
Query 91 HKWKASLLYILGVLLIALGYSLIGLPLQLYGLLRLFASFLPQVLGAVRLSPIGCWILQLP 150
+K ++ + +G + +G+ ++G+ L+ YG LF+ F P + + P+ WI+Q P
Sbjct 65 QNYKGTISFGVGFFFVIIGWPILGMMLETYGFFVLFSGFWPTLAVFAQKIPVLGWIIQQP 124
Query 151 GLKQ 154
++
Sbjct 125 YIRS 128
> At3g49420
Length=140
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 76/124 (61%), Gaps = 1/124 (0%)
Query 31 LSDGQRIGVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKP 90
++D ++IG+GL G+ F+G++ D+ LL +GN+ F+SG+ L +G + T++FF +K
Sbjct 6 MNDRKKIGLGLTGFGVFFSFLGIVFVFDKGLLAMGNILFISGVSLTIGFKSTMQFF-MKR 64
Query 91 HKWKASLLYILGVLLIALGYSLIGLPLQLYGLLRLFASFLPQVLGAVRLSPIGCWILQLP 150
+K ++ + +G + +G+ ++G+ L+ YG LF+ F P + + P+ WI+Q P
Sbjct 65 QNYKGTISFGVGFFFVIIGWPILGMMLETYGFFVLFSGFWPTLAVFAQKIPVLGWIIQQP 124
Query 151 GLKQ 154
++
Sbjct 125 YIRS 128
> CE02732
Length=112
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 40/100 (40%), Positives = 61/100 (61%), Gaps = 1/100 (1%)
Query 56 FLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKPHKWKASLLYILGVLLIALGYSLIGL 115
FLD ALL +GNL F+ GI ++G ++TL FF + K K S+L+ G+L++ GY L G+
Sbjct 2 FLDSALLAIGNLLFIVGITFIIGVQRTLVFF-FEFRKLKGSILFFGGILVVLFGYPLFGM 60
Query 116 PLQLYGLLRLFASFLPQVLGAVRLSPIGCWILQLPGLKQC 155
+ +G + LF FLP ++ +R P I LPG++Q
Sbjct 61 IAECWGFIVLFGGFLPGIVNLLRSIPGISTITYLPGIRQV 100
> YMR292w
Length=138
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 46/138 (33%), Positives = 76/138 (55%), Gaps = 2/138 (1%)
Query 31 LSDGQRIGVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKP 90
L++ Q+ GV G G+ F DRALL LGN+ F+ G++L++G +KT FF +P
Sbjct 3 LTEAQKFGVAFTFGGFLFFLFGIFTFFDRALLALGNILFLIGVFLIIGSQKTYIFFT-RP 61
Query 91 HKWKASLLYILGVLLIALGYSLIGLPLQLYGLLRLFASFLPQVLGAVRLSPIGCWILQLP 150
+K + SL +++G LI L ++ +G ++ G++ LF F ++ +R PI IL P
Sbjct 62 NKRRGSLFFLVGAFLILLKWTFLGFIIESLGIIGLFGDFFGVIVQFLRSMPIIGPILSHP 121
Query 151 GLKQCTKWVLDGDKQLPL 168
+ L G + LP+
Sbjct 122 AIAPIVDK-LAGVRVLPV 138
> Hs17488855
Length=126
Score = 62.4 bits (150), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 38/101 (37%), Positives = 56/101 (55%), Gaps = 1/101 (0%)
Query 35 QRIGVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKPHKWK 94
IGVG+ G+ G L + D LL GNL F++G+ L++G RKT FF + HK K
Sbjct 2 AEIGVGITGFGIFFILFGTLLYFDSVLLAFGNLLFLTGLSLIIGLRKTFWFF-FQRHKLK 60
Query 95 ASLLYILGVLLIALGYSLIGLPLQLYGLLRLFASFLPQVLG 135
+ + GV+++ L + L+G+ L+ YG LF F P G
Sbjct 61 GTSFLLGGVVIVLLRWPLLGMFLETYGFFSLFKGFFPVAFG 101
> At3g03180
Length=122
Score = 61.2 bits (147), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 60/96 (62%), Gaps = 1/96 (1%)
Query 31 LSDGQRIGVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKP 90
++D ++IG+GL G+ F+G++ D+ L+ +GN+ F++G+ L +G ++FF K
Sbjct 6 MNDLKKIGLGLTGFGVFFTFLGVIFVFDKGLIAMGNILFLAGVTLTIGINPAIQFFT-KR 64
Query 91 HKWKASLLYILGVLLIALGYSLIGLPLQLYGLLRLF 126
+K ++ + LG LL+ G+ + GL L+ YG L LF
Sbjct 65 QNFKGTISFGLGFLLVVFGWPIFGLLLESYGFLVLF 100
> SPCC4B3.02c
Length=129
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 68/126 (53%), Gaps = 2/126 (1%)
Query 31 LSDGQRIGVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKP 90
LSD Q+IGVG LG +G+ F D LL+LGNL + G +++ G K++ FF L+
Sbjct 3 LSDLQKIGVGTTALGFLFMIMGIFMFFDGPLLSLGNLLLVFGFFMIAGFSKSVSFF-LRK 61
Query 91 HKWKASLLYILGVLLIALGYSLIGLPLQLYGLLRLFASFLPQVLGAVRLSP-IGCWILQL 149
+ S+ + G+LL + +IG ++ G LF F P ++ +R P IG +I +L
Sbjct 62 DRMLGSISFFSGLLLTLFHFPIIGFFVECLGFFNLFKVFYPLIISFLRTVPYIGPYIDRL 121
Query 150 PGLKQC 155
+Q
Sbjct 122 TSYQQS 127
> ECU10g0860
Length=117
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 44/77 (57%), Gaps = 3/77 (3%)
Query 38 GVGLCLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLVLGGRKTLKFFVLKPHKWKASL 97
GV + +G+S+ VG + +DRAL+ GNL + GI L+ R F +L+P K + +
Sbjct 7 GVVVVSIGMSVFLVGAILLMDRALMISGNLLMIIGISLLARSR---MFALLRPEKIQGMV 63
Query 98 LYILGVLLIALGYSLIG 114
++ +GVL + + + G
Sbjct 64 IFAMGVLFLIYRFPMFG 80
> CE08772
Length=708
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 35/62 (56%), Gaps = 9/62 (14%)
Query 70 MSGIWLVLGGRKTLKFFVLKPHKWKASLLYILGVLLIALGYSLI-GLPLQLYGLLRLFAS 128
++G W+ L GR++L+F K W++ L IA+G++LI + +++Y L+ F
Sbjct 615 INGFWVALVGRQSLQFAFTKYRFWES--------LGIAIGFALIRHVTVEIYLLITFFML 666
Query 129 FL 130
L
Sbjct 667 LL 668
> At4g03490
Length=587
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 28/95 (29%), Positives = 47/95 (49%), Gaps = 13/95 (13%)
Query 21 CLTEPRFRAMLSDGQRIGVGL----CLLGLSLGFVGMLCFLDRALLTLGNLAFMSGIWLV 76
C + + A++S IG L C+L V +L F A+ +LAF++G++LV
Sbjct 402 CNSIAVYTAVISTVALIGTQLADLKCMLTTFKFIVPLLGFSIIAM----SLAFVAGLYLV 457
Query 77 LGGRKTLKFFVLKPHKWKASLLYILGVLLIALGYS 111
LG L FVL + Y++ +LL+ + Y+
Sbjct 458 LGHHYWLAIFVL-----ASGGFYLMALLLLIIPYA 487
Lambda K H
0.331 0.150 0.494
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2418402922
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40