bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1749_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
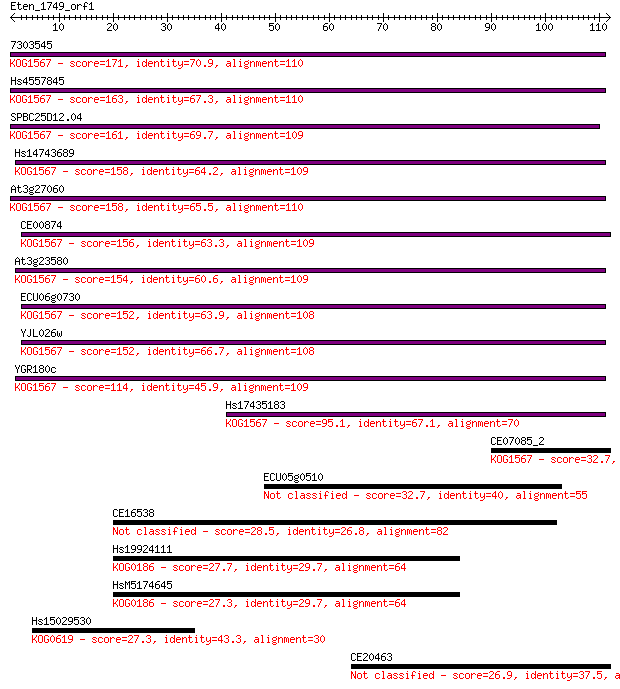
7303545 171 4e-43
Hs4557845 163 9e-41
SPBC25D12.04 161 3e-40
Hs14743689 158 2e-39
At3g27060 158 3e-39
CE00874 156 9e-39
At3g23580 154 5e-38
ECU06g0730 152 1e-37
YJL026w 152 2e-37
YGR180c 114 5e-26
Hs17435183 95.1 3e-20
CE07085_2 32.7 0.15
ECU05g0510 32.7 0.16
CE16538 28.5 3.5
Hs19924111 27.7 5.5
HsM5174645 27.3 6.8
Hs15029530 27.3 7.3
CE20463 26.9 9.2
> 7303545
Length=383
Score = 171 bits (432), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 78/110 (70%), Positives = 90/110 (81%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
PRR+V+ PI +H++W MYKKAEASFWT EE+D+S DL DW L +E+HFI HVLAFFAA
Sbjct 71 PRRFVIFPIQYHDIWQMYKKAEASFWTVEEVDLSKDLTDWHRLKDDERHFISHVLAFFAA 130
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQI EAR FY FQIA+EN+HSE YSVLID YIR+
Sbjct 131 SDGIVNENLVERFSQEVQITEARCFYGFQIAMENVHSEMYSVLIDTYIRD 180
> Hs4557845
Length=389
Score = 163 bits (412), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 74/110 (67%), Positives = 89/110 (80%), Gaps = 0/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAA 60
PRR+V+ PI +H++W MYKKAEASFWT EE+D+S D+ W +L E++FI HVLAFFAA
Sbjct 77 PRRFVIFPIEYHDIWQMYKKAEASFWTAEEVDLSKDIQHWESLKPEERYFISHVLAFFAA 136
Query 61 SDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
SDGIV ENL ERF EVQI EAR FY FQIA+ENIHSE YS+LID YI++
Sbjct 137 SDGIVNENLVERFSQEVQITEARCFYGFQIAMENIHSEMYSLLIDTYIKD 186
> SPBC25D12.04
Length=391
Score = 161 bits (407), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 76/110 (69%), Positives = 90/110 (81%), Gaps = 1/110 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWAT-LDANEQHFIKHVLAFFA 59
P R+V+ PI +HE+W YKKAEASFWT EEID+S DLVDW L+A+E++FI VLA+FA
Sbjct 73 PHRFVLFPIKYHEIWQFYKKAEASFWTAEEIDLSKDLVDWDNKLNADERYFISTVLAYFA 132
Query 60 ASDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIR 109
ASDGIV ENL ERF +EVQIPEAR Y FQI +ENIHSETYS+L+D YIR
Sbjct 133 ASDGIVNENLLERFSSEVQIPEARCVYGFQIMIENIHSETYSLLLDTYIR 182
> Hs14743689
Length=351
Score = 158 bits (400), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 70/109 (64%), Positives = 88/109 (80%), Gaps = 0/109 (0%)
Query 2 RRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAAS 61
RR+V+ PI + ++W MYK+A+ASFWT EE+D+S DL W L A+E++FI H+LAFFAAS
Sbjct 40 RRFVIFPIQYPDIWKMYKQAQASFWTAEEVDLSKDLPHWNKLKADEKYFISHILAFFAAS 99
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
DGIV ENL ERF EVQ+PEAR FY FQI +EN+HSE YS+LID YIR+
Sbjct 100 DGIVNENLVERFSQEVQVPEARCFYGFQILIENVHSEMYSLLIDTYIRD 148
> At3g27060
Length=332
Score = 158 bits (399), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 72/111 (64%), Positives = 92/111 (82%), Gaps = 1/111 (0%)
Query 1 PRRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWA-TLDANEQHFIKHVLAFFA 59
P R+ + PI + ++W MYKKAEASFWT EE+D+S D DW +L+ E+HFIKHVLAFFA
Sbjct 14 PDRFCMFPIHYPQIWEMYKKAEASFWTAEEVDLSQDNRDWENSLNDGERHFIKHVLAFFA 73
Query 60 ASDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
ASDGIV+ENLA RFM++VQ+ EAR+FY FQIA+ENIHSE YS+L+D YI++
Sbjct 74 ASDGIVLENLASRFMSDVQVSEARAFYGFQIAIENIHSEMYSLLLDTYIKD 124
> CE00874
Length=381
Score = 156 bits (394), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 69/109 (63%), Positives = 86/109 (78%), Gaps = 0/109 (0%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
R+V+ P+ HH++WN YKKA ASFWT EE+D+ D+ DW ++ +EQ+FI +LAFFAASD
Sbjct 71 RFVIFPLKHHDIWNFYKKAVASFWTVEEVDLGKDMNDWEKMNGDEQYFISRILAFFAASD 130
Query 63 GIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRNE 111
GIV ENL ERF EVQ+ EAR FY FQIA+ENIHSE YS LI+ YIR+E
Sbjct 131 GIVNENLCERFSNEVQVSEARFFYGFQIAIENIHSEMYSKLIETYIRDE 179
> At3g23580
Length=341
Score = 154 bits (388), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 66/109 (60%), Positives = 89/109 (81%), Gaps = 0/109 (0%)
Query 2 RRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAAS 61
+R+ + PI + +W MYKKAEASFWT EE+D+S D+ W L +E+HFI H+LAFFAAS
Sbjct 27 QRFTMFPIRYKSIWEMYKKAEASFWTAEEVDLSTDVQQWEALTDSEKHFISHILAFFAAS 86
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
DGIV+ENLA RF+ +VQ+PEAR+FY FQIA+ENIHSE YS+L++ +I++
Sbjct 87 DGIVLENLAARFLNDVQVPEARAFYGFQIAMENIHSEMYSLLLETFIKD 135
> ECU06g0730
Length=325
Score = 152 bits (385), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 69/108 (63%), Positives = 87/108 (80%), Gaps = 0/108 (0%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASD 62
R+V+LPI +H++W MYKKAE+SFWT EE+ + D+ DW L+A E+HFI +VLAFFAASD
Sbjct 17 RFVLLPIKYHDIWKMYKKAESSFWTVEEVSLDKDIDDWGKLNAKERHFISYVLAFFAASD 76
Query 63 GIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
GIV NL ERF EV++ EAR FY FQ+A+ENIHSE YS+LID YIR+
Sbjct 77 GIVNLNLVERFSTEVKVLEARFFYGFQMAIENIHSEMYSLLIDTYIRD 124
> YJL026w
Length=399
Score = 152 bits (383), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 72/109 (66%), Positives = 86/109 (78%), Gaps = 1/109 (0%)
Query 3 RWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWAT-LDANEQHFIKHVLAFFAAS 61
R V+ PI +HE+W YK+AEASFWT EEID+S D+ DW ++ NE+ FI VLAFFAAS
Sbjct 85 RTVLFPIKYHEIWQAYKRAEASFWTAEEIDLSKDIHDWNNRMNENERFFISRVLAFFAAS 144
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
DGIV ENL E F EVQIPEA+SFY FQI +ENIHSETYS+LID YI++
Sbjct 145 DGIVNENLVENFSTEVQIPEAKSFYGFQIMIENIHSETYSLLIDTYIKD 193
> YGR180c
Length=345
Score = 114 bits (284), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 50/109 (45%), Positives = 78/109 (71%), Gaps = 0/109 (0%)
Query 2 RRWVVLPIVHHEVWNMYKKAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAAS 61
RR+V+ PI +HE+W YKK EASFWT EEI+++ D D+ L +++ +I ++LA +S
Sbjct 33 RRFVMFPIKYHEIWAAYKKVEASFWTAEEIELAKDTEDFQKLTDDQKTYIGNLLALSISS 92
Query 62 DGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETYSVLIDLYIRN 110
D +V + L E F A++Q PE +SFY FQI +ENI+SE YS+++D + ++
Sbjct 93 DNLVNKYLIENFSAQLQNPEGKSFYGFQIMMENIYSEVYSMMVDAFFKD 141
> Hs17435183
Length=213
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 47/70 (67%), Positives = 52/70 (74%), Gaps = 0/70 (0%)
Query 41 ATLDANEQHFIKHVLAFFAASDGIVMENLAERFMAEVQIPEARSFYAFQIAVENIHSETY 100
A + E FI VLAFFAASDGIV ENL ERF EVQI EAR FY FQIA+EN HSE Y
Sbjct 64 APGETEESDFISRVLAFFAASDGIVNENLVERFSQEVQITEARCFYDFQIAMENTHSEMY 123
Query 101 SVLIDLYIRN 110
S+LID YI++
Sbjct 124 SLLIDTYIKD 133
> CE07085_2
Length=224
Score = 32.7 bits (73), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 14/22 (63%), Positives = 19/22 (86%), Gaps = 0/22 (0%)
Query 90 IAVENIHSETYSVLIDLYIRNE 111
IAVENIHSE Y+ L++ YIR++
Sbjct 1 IAVENIHSEMYAKLLEAYIRDD 22
> ECU05g0510
Length=230
Score = 32.7 bits (73), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 34/64 (53%), Gaps = 10/64 (15%)
Query 48 QHFIKHVLAFF--AASDGIVMENLAERFM-------AEVQIPEARSFYAFQIAVENIHSE 98
+++I+H+ +GIV E L RF+ E QIPE SF +Q+ V+N ++
Sbjct 39 RYYIEHLGCRMRRKGENGIVPE-LICRFLVDEEYGECEYQIPELLSFLRYQVTVKNYLND 97
Query 99 TYSV 102
YSV
Sbjct 98 RYSV 101
> CE16538
Length=129
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 35/82 (42%), Gaps = 9/82 (10%)
Query 20 KAEASFWTTEEIDMSADLVDWATLDANEQHFIKHVLAFFAASDGIVMENLAERFMAEVQI 79
KAEA++WT ++ + DW H L +A D IVM + ++ + +
Sbjct 56 KAEATYWTGLDLHKGETMSDWTN-----THLRGDQLR-YAIMDTIVMHLIIKQRLRNIHK 109
Query 80 PEARSFYAFQIAVENIHSETYS 101
+ R FQ EN H Y+
Sbjct 110 NDPR---LFQYVFENGHPLPYN 128
> Hs19924111
Length=600
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 5/67 (7%)
Query 20 KAEASFWTTEE---IDMSADLVDWATLDANEQHFIKHVLAFFAASDGIVMENLAERFMAE 76
+AE W T E + + DL+DW++L + KH++ A + +E L RF E
Sbjct 294 RAEIEDWFTAETLGVSGTMDLLDWSSLIDSRTKLSKHLVVPNAQTGQ--LEPLLSRFTEE 351
Query 77 VQIPEAR 83
++ R
Sbjct 352 EELQMTR 358
> HsM5174645
Length=516
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 5/67 (7%)
Query 20 KAEASFWTTEE---IDMSADLVDWATLDANEQHFIKHVLAFFAASDGIVMENLAERFMAE 76
+AE W T E + + DL+DW++L + KH++ A + +E L RF E
Sbjct 210 RAEIEDWFTAETLGVSGTMDLLDWSSLIDSRTKLSKHLVVPNAQTGQ--LEPLLSRFTEE 267
Query 77 VQIPEAR 83
++ R
Sbjct 268 EELQMTR 274
> Hs15029530
Length=653
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 1/31 (3%)
Query 5 VVLPIVH-HEVWNMYKKAEASFWTTEEIDMS 34
VVLP +H H +N YK A + WT + S
Sbjct 596 VVLPTIHDHINYNTYKPAHGAHWTENSLGNS 626
> CE20463
Length=1093
Score = 26.9 bits (58), Expect = 9.2, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 64 IVMENLAERFMAEVQIPE---ARSFYAFQIAVENIHSET-YSVLIDLYIRNE 111
I +NL + ++PE AR AF + +E IHSET + L+ Y+ E
Sbjct 983 ISAKNLTIEHIYHSEMPEKHSARRAIAFGVKIEEIHSETEFEKLLSEYVEYE 1034
Lambda K H
0.323 0.133 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40