bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
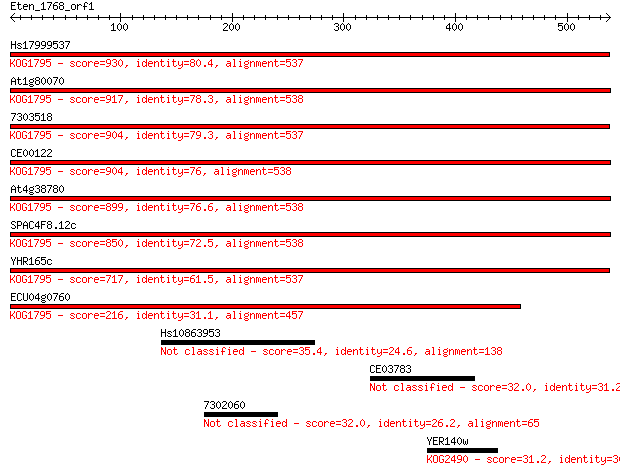
Query= Eten_1768_orf1
Length=538
Score E
Sequences producing significant alignments: (Bits) Value
Hs17999537 930 0.0
At1g80070 917 0.0
7303518 904 0.0
CE00122 904 0.0
At4g38780 899 0.0
SPAC4F8.12c 850 0.0
YHR165c 717 0.0
ECU04g0760 216 1e-55
Hs10863953 35.4 0.30
CE03783 32.0 3.3
7302060 32.0 3.5
YER140w 31.2 6.7
> Hs17999537
Length=2335
Score = 930 bits (2403), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 432/537 (80%), Positives = 493/537 (91%), Gaps = 0/537 (0%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HESIVMDLCQVFD ELD+LE+ETVQKETIHPRKSYKMNSSCADILLFA+YKW++S+PSL
Sbjct 1585 IHESIVMDLCQVFDQELDALEIETVQKETIHPRKSYKMNSSCADILLFASYKWNVSRPSL 1644
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
LA+ KD+M+ T K+W+D+QLRWGDYDSHDIERY R+KFLDYTTDNMSIYPSP+GVL+
Sbjct 1645 LADSKDVMDSTTTQKYWIDIQLRWGDYDSHDIERYARAKFLDYTTDNMSIYPSPTGVLIA 1704
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DLAYNLHSA+GNW PG KPL+Q+AM KIMKANPALYVLRERIRKGLQLYSSEPTEPYL+
Sbjct 1705 IDLAYNLHSAYGNWFPGSKPLIQQAMAKIMKANPALYVLRERIRKGLQLYSSEPTEPYLS 1764
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
SQNYGELFSNQ +WF+DDTNVYRVTIHKTFEGNLTTKP+NG IFIFNPRTGQLFLKIIHT
Sbjct 1765 SQNYGELFSNQIIWFVDDTNVYRVTIHKTFEGNLTTKPINGAIFIFNPRTGQLFLKIIHT 1824
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
SVWAGQKRL QLA+WKTAEEVAALIRSLPVEEQPKQII TRKGMLDPLEVHLLDFPNIVI
Sbjct 1825 SVWAGQKRLGQLAKWKTAEEVAALIRSLPVEEQPKQIIVTRKGMLDPLEVHLLDFPNIVI 1884
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
KGSEL LPFQA +++EKFGD+IL+AT+P+MVLFN++DDWLK+IS+YTAFSRL+L+LRAL
Sbjct 1885 KGSELQLPFQACLKVEKFGDLILKATEPQMVLFNLYDDWLKTISSYTAFSRLILILRALH 1944
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
VN +RAKV+L+P K+ +T+PHHIWP+LTDEEWI VEV LKDLIL DY KKNNVNVASLTQ
Sbjct 1945 VNNDRAKVILKPDKTTITEPHHIWPTLTDEEWIKVEVQLKDLILADYGKKNNVNVASLTQ 2004
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQVTSTTTRSVNIHGEEMIVATQSPHEQ 480
SEIRDIILGMEI+ PS QRQQIAEIE Q +EQ Q+T+T TR+VN HG+E+I +T S +E
Sbjct 2005 SEIRDIILGMEISAPSQQRQQIAEIEKQTKEQSQLTATQTRTVNKHGDEIITSTTSNYET 2064
Query 481 QVFSSKTDWRVRAISAASLHLRTRHLYVSSEAVAPAGCTYILPKNLLKKFISIADLR 537
Q FSSKT+WRVRAISAA+LHLRT H+YVSS+ + G TYILPKN+LKKFI I+DLR
Sbjct 2065 QTFSSKTEWRVRAISAANLHLRTNHIYVSSDDIKETGYTYILPKNVLKKFICISDLR 2121
> At1g80070
Length=2382
Score = 917 bits (2371), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 421/538 (78%), Positives = 489/538 (90%), Gaps = 0/538 (0%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HES+VMDLCQV D ELD+LE+ETVQKETIHPRKSYKMNSSCAD+LLFA +KW +SKPSL
Sbjct 1632 IHESVVMDLCQVLDQELDALEIETVQKETIHPRKSYKMNSSCADVLLFAAHKWPMSKPSL 1691
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
+AE KD+ + + K+W+DVQLRWGDYDSHDIERY R+KF+DYTTDNMSIYPSP+GV++G
Sbjct 1692 VAESKDMFDQKASNKYWIDVQLRWGDYDSHDIERYTRAKFMDYTTDNMSIYPSPTGVMIG 1751
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DLAYNLHSAFGNW PG KPL+ +AMNKIMK+NPALYVLRERIRKGLQLYSSEPTEPYL+
Sbjct 1752 LDLAYNLHSAFGNWFPGSKPLLAQAMNKIMKSNPALYVLRERIRKGLQLYSSEPTEPYLS 1811
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
SQNYGE+FSNQ +WF+DDTNVYRVTIHKTFEGNLTTKP+NG IFIFNPRTGQLFLK+IHT
Sbjct 1812 SQNYGEIFSNQIIWFVDDTNVYRVTIHKTFEGNLTTKPINGAIFIFNPRTGQLFLKVIHT 1871
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
SVWAGQKRL QLA+WKTAEEVAAL+RSLPVEEQPKQII TRKGMLDPLEVHLLDFPNIVI
Sbjct 1872 SVWAGQKRLGQLAKWKTAEEVAALVRSLPVEEQPKQIIVTRKGMLDPLEVHLLDFPNIVI 1931
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
KGSEL LPFQA ++IEKFGD+IL+AT+P+MVLFN++DDWLKSIS+YTAFSRL+L+LRAL
Sbjct 1932 KGSELQLPFQACLKIEKFGDLILKATEPQMVLFNIYDDWLKSISSYTAFSRLILILRALH 1991
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
VN E+AK+LL+P KSVVT+PHHIWPSLTD++W+ VEV L+DLIL DYAKKNNVN ++LTQ
Sbjct 1992 VNNEKAKMLLKPDKSVVTEPHHIWPSLTDDQWMKVEVALRDLILSDYAKKNNVNTSALTQ 2051
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQVTSTTTRSVNIHGEEMIVATQSPHEQ 480
SEIRDIILG EI PPS QRQQIAEIE QA+E Q+T+ TTR+ N+HG+E+IV T SP+EQ
Sbjct 2052 SEIRDIILGAEITPPSQQRQQIAEIEKQAKEASQLTAVTTRTTNVHGDELIVTTTSPYEQ 2111
Query 481 QVFSSKTDWRVRAISAASLHLRTRHLYVSSEAVAPAGCTYILPKNLLKKFISIADLRT 538
F SKTDWRVRAISA +L+LR H+YV+S+ + G TYI+PKN+LKKFI +ADLRT
Sbjct 2112 SAFGSKTDWRVRAISATNLYLRVNHIYVNSDDIKETGYTYIMPKNILKKFICVADLRT 2169
> 7303518
Length=2396
Score = 904 bits (2336), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 426/537 (79%), Positives = 487/537 (90%), Gaps = 0/537 (0%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HESIVMDLCQVFD ELD+LE+ETVQKETIHPRKSYKMNSSCADILLF YKW++S+PSL
Sbjct 1645 IHESIVMDLCQVFDQELDALEIETVQKETIHPRKSYKMNSSCADILLFPAYKWNVSRPSL 1704
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
LA+ KD M+ T K+WLD+QLRWGDYDSHD+ERY R+KFLDYTTDNMSIYPSP+GVL+
Sbjct 1705 LADTKDTMDNTTTQKYWLDIQLRWGDYDSHDVERYARAKFLDYTTDNMSIYPSPTGVLIA 1764
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DLAYNLHSA+GNW PG K L+Q+AM KIMKANPALYVLRERIRK LQLYSSEPTEPYL+
Sbjct 1765 IDLAYNLHSAYGNWFPGCKTLIQQAMAKIMKANPALYVLRERIRKALQLYSSEPTEPYLS 1824
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
SQNYGELFSNQ +WF+DDTNVYRVTIHKTFEGNLTTKP+NG IFIFNPRTGQLFLKIIHT
Sbjct 1825 SQNYGELFSNQIIWFVDDTNVYRVTIHKTFEGNLTTKPINGAIFIFNPRTGQLFLKIIHT 1884
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
SVWAGQKRL QLA+WKTAEEVAALIRSLPVEEQPKQII TRKGMLDPLEVHLLDFPNIVI
Sbjct 1885 SVWAGQKRLGQLAKWKTAEEVAALIRSLPVEEQPKQIIVTRKGMLDPLEVHLLDFPNIVI 1944
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
KGSEL LPFQA +++EKFGD+IL+AT+P+MVLFN++DDWLK+IS+YTAFSRL+L+LRAL
Sbjct 1945 KGSELQLPFQACLKVEKFGDLILKATEPQMVLFNLYDDWLKTISSYTAFSRLILILRALH 2004
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
VNTER K++L+P K+ +T+ HHIWP+LTDEEWI VEV LKDLIL DY KKNNVNVASLTQ
Sbjct 2005 VNTERTKIILKPDKTTITEAHHIWPTLTDEEWIKVEVQLKDLILADYGKKNNVNVASLTQ 2064
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQVTSTTTRSVNIHGEEMIVATQSPHEQ 480
SEIRDIILGMEI+ PS QRQQIAEIE Q +EQ Q+T+TTTR+ N HG+E+I +T S +E
Sbjct 2065 SEIRDIILGMEISAPSAQRQQIAEIEKQTKEQNQLTATTTRTTNKHGDEIITSTTSNYET 2124
Query 481 QVFSSKTDWRVRAISAASLHLRTRHLYVSSEAVAPAGCTYILPKNLLKKFISIADLR 537
Q FSSKT+WRVRAISA +LHLRT H+YVSS+ + G TYILPKN+LKKF++I+DLR
Sbjct 2125 QTFSSKTEWRVRAISATNLHLRTNHIYVSSDDIKETGYTYILPKNILKKFVTISDLR 2181
> CE00122
Length=2329
Score = 904 bits (2335), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 409/538 (76%), Positives = 491/538 (91%), Gaps = 0/538 (0%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HES+VMDLCQVFD ELD+LE++TVQKETIHPRKSYKMNSSCAD+LLFA YKW++S+PSL
Sbjct 1578 IHESVVMDLCQVFDQELDALEIQTVQKETIHPRKSYKMNSSCADVLLFAQYKWNVSRPSL 1637
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
+A+ KD+M+ T K+WLDVQLRWGDYDSHD+ERY R+KFLDYTTDNMSIYPSP+GVL+
Sbjct 1638 MADSKDVMDNTTTQKYWLDVQLRWGDYDSHDVERYARAKFLDYTTDNMSIYPSPTGVLIA 1697
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DLAYNL+SA+GNW PG+KPL+++AM KI+KANPA YVLRERIRKGLQLYSSEPTEPYL
Sbjct 1698 IDLAYNLYSAYGNWFPGMKPLIRQAMAKIIKANPAFYVLRERIRKGLQLYSSEPTEPYLT 1757
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
SQNYGELFSNQ +WF+DDTNVYRVTIHKTFEGNLTTKP+NG IFIFNPRTGQLFLKIIHT
Sbjct 1758 SQNYGELFSNQIIWFVDDTNVYRVTIHKTFEGNLTTKPINGAIFIFNPRTGQLFLKIIHT 1817
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
SVWAGQKRL QLA+WKTAEEVAALIRSLPVEEQP+QII TRK MLDPLEVHLLDFPNIVI
Sbjct 1818 SVWAGQKRLSQLAKWKTAEEVAALIRSLPVEEQPRQIIVTRKAMLDPLEVHLLDFPNIVI 1877
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
KGSEL LPFQA+M++EKFGD+IL+AT+P+MVLFN++DDWLK+IS+YTAFSR++L++R +
Sbjct 1878 KGSELMLPFQAIMKVEKFGDLILKATEPQMVLFNLYDDWLKTISSYTAFSRVVLIMRGMH 1937
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
+N ++ KV+L+P K+ +T+PHHIWP+L+D++WI VE+ LKD+IL DY KKNNVNVASLTQ
Sbjct 1938 INPDKTKVILKPDKTTITEPHHIWPTLSDDDWIKVELALKDMILADYGKKNNVNVASLTQ 1997
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQVTSTTTRSVNIHGEEMIVATQSPHEQ 480
SE+RDIILGMEI+ PS QRQQIA+IE Q +EQ QVT+TTTR+VN HG+E+I AT S +E
Sbjct 1998 SEVRDIILGMEISAPSQQRQQIADIEKQTKEQSQVTATTTRTVNKHGDEIITATTSNYET 2057
Query 481 QVFSSKTDWRVRAISAASLHLRTRHLYVSSEAVAPAGCTYILPKNLLKKFISIADLRT 538
F+S+T+WRVRAIS+ +LHLRT+H+YV+S+ V G TYILPKN+LKKFI+I+DLRT
Sbjct 2058 ASFASRTEWRVRAISSTNLHLRTQHIYVNSDDVKDTGYTYILPKNILKKFITISDLRT 2115
> At4g38780
Length=2352
Score = 899 bits (2323), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 412/538 (76%), Positives = 483/538 (89%), Gaps = 0/538 (0%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HES+VMDLCQV D EL+ LE+ETVQKETIHPRKSYKMNSSCAD+LLFA +KW +SKPSL
Sbjct 1604 IHESVVMDLCQVLDQELEPLEIETVQKETIHPRKSYKMNSSCADVLLFAAHKWPMSKPSL 1663
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
+AE KD+ + + K+W+DVQLRWGDYDSHDIERY ++KF+DYTTDNMSIYPSP+GV++G
Sbjct 1664 IAESKDVFDQKASNKYWIDVQLRWGDYDSHDIERYTKAKFMDYTTDNMSIYPSPTGVIIG 1723
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DLAYNLHSAFGNW PG KPL+ +AMNKIMK+NPALYVLRERIRKGLQLYSSEPTEPYL+
Sbjct 1724 LDLAYNLHSAFGNWFPGSKPLLAQAMNKIMKSNPALYVLRERIRKGLQLYSSEPTEPYLS 1783
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
SQNYGE+FSNQ +WF+DDTNVYRVTIHKTFEGNLTTKP+NGVIFIFNPRTGQLFLKIIHT
Sbjct 1784 SQNYGEIFSNQIIWFVDDTNVYRVTIHKTFEGNLTTKPINGVIFIFNPRTGQLFLKIIHT 1843
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
SVWAGQKRL QLA+WKTAEEVAAL+RSLPVEEQPKQ+I TRKGMLDPLEVHLLDFPNIVI
Sbjct 1844 SVWAGQKRLGQLAKWKTAEEVAALVRSLPVEEQPKQVIVTRKGMLDPLEVHLLDFPNIVI 1903
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
KGSEL LPFQA ++IEKFGD+IL+AT+P+M LFN++DDWL ++S+YTAF RL+L+LRAL
Sbjct 1904 KGSELQLPFQACLKIEKFGDLILKATEPQMALFNIYDDWLMTVSSYTAFQRLILILRALH 1963
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
VN E+AK+LL+P SVVT+P+HIWPSLTD++W+ VEV L+DLIL DYAKKN VN ++LTQ
Sbjct 1964 VNNEKAKMLLKPDMSVVTEPNHIWPSLTDDQWMKVEVALRDLILSDYAKKNKVNTSALTQ 2023
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQVTSTTTRSVNIHGEEMIVATQSPHEQ 480
SEIRDIILG EI PPS QRQQIAEIE QA+E Q+T+ TTR+ N+HG+E+I T SP+EQ
Sbjct 2024 SEIRDIILGAEITPPSQQRQQIAEIEKQAKEASQLTAVTTRTTNVHGDELISTTISPYEQ 2083
Query 481 QVFSSKTDWRVRAISAASLHLRTRHLYVSSEAVAPAGCTYILPKNLLKKFISIADLRT 538
F SKTDWRVRAISA +L+LR H+YV+S+ + G TYI+PKN+LKKFI IADLRT
Sbjct 2084 SAFGSKTDWRVRAISATNLYLRVNHIYVNSDDIKETGYTYIMPKNILKKFICIADLRT 2141
> SPAC4F8.12c
Length=2363
Score = 850 bits (2197), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 390/539 (72%), Positives = 477/539 (88%), Gaps = 1/539 (0%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HES+V DLCQV D EL+SL++ETVQKETIHPRKSYKMNSSCADILL A YKW++S+PSL
Sbjct 1609 IHESVVWDLCQVLDQELESLQIETVQKETIHPRKSYKMNSSCADILLLAAYKWNVSRPSL 1668
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
L + +D+++ T K+W+DVQLR+GDYDSHDIERY R+KFLDY+TD S+YPSP+GVL+G
Sbjct 1669 LNDNRDVLDNTTTNKYWIDVQLRFGDYDSHDIERYTRAKFLDYSTDAQSMYPSPTGVLIG 1728
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DL YN+HSA+GNWIPG+KPL+Q++MNKIMKANPALYVLRERIRKGLQLY+SEP E YL+
Sbjct 1729 IDLCYNMHSAYGNWIPGMKPLIQQSMNKIMKANPALYVLRERIRKGLQLYASEPQEQYLS 1788
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
S NY ELFSNQ F+DDTNVYRVTIHKTFEGNLTTKP+NG IFIFNPRTGQLFLK+IHT
Sbjct 1789 SSNYAELFSNQIQLFVDDTNVYRVTIHKTFEGNLTTKPINGAIFIFNPRTGQLFLKVIHT 1848
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
SVWAGQKRL QLA+WKTAEEVAALIRSLPVEEQP+QII TRKGMLDPLEVHLLDFPNI I
Sbjct 1849 SVWAGQKRLGQLAKWKTAEEVAALIRSLPVEEQPRQIIVTRKGMLDPLEVHLLDFPNITI 1908
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
KGSEL LPFQA+++++K D+ILRAT+P+MVLFN++DDWL+S+S+YTAFSRL+L+LRAL
Sbjct 1909 KGSELQLPFQAIIKLDKINDLILRATEPQMVLFNLYDDWLQSVSSYTAFSRLILILRALN 1968
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
VNTE+ K++LRP KS++T+ +H+WP+L D++W+ VE L+DLIL DYAKKNN+NVASLT
Sbjct 1969 VNTEKTKLILRPDKSIITKENHVWPNLDDQQWLDVEPKLRDLILADYAKKNNINVASLTN 2028
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQVTSTTTRSVNIHGEEMIVATQSPHEQ 480
SE+RDIILGM I PSLQRQQIAEIE Q RE QVT+ TT++ N+HG+EM+V T S +E
Sbjct 2029 SEVRDIILGMTITAPSLQRQQIAEIEKQGRENAQVTAVTTKTTNVHGDEMVVTTTSAYEN 2088
Query 481 QVFSSKTDWRVRAISAASLHLRTRHLYVSSEAVAPA-GCTYILPKNLLKKFISIADLRT 538
+ FSSKT+WR RAIS+ SL LRT+++YV+S+ ++ TYILP+NLL+KF++I+DLRT
Sbjct 2089 EKFSSKTEWRNRAISSISLPLRTKNIYVNSDNISETFPYTYILPQNLLRKFVTISDLRT 2147
> YHR165c
Length=2413
Score = 717 bits (1852), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 330/544 (60%), Positives = 424/544 (77%), Gaps = 7/544 (1%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HESIV D+CQ+ D ELD L++E+V KET+HPRKSYKMNSS ADI + + ++W +SKPSL
Sbjct 1657 IHESIVFDICQILDGELDVLQIESVTKETVHPRKSYKMNSSAADITMESVHEWEVSKPSL 1716
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
L E D +G K W DVQLR+GDYDSHDI RY R+KFLDYTTDN+S+YPSP+GV++G
Sbjct 1717 LHETNDSFKGLITNKMWFDVQLRYGDYDSHDISRYVRAKFLDYTTDNVSMYPSPTGVMIG 1776
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DLAYN++ A+GNW GLKPL+Q +M IMKANPALYVLRERIRKGLQ+Y S EP+LN
Sbjct 1777 IDLAYNMYDAYGNWFNGLKPLIQNSMRTIMKANPALYVLRERIRKGLQIYQSSVQEPFLN 1836
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
S NY ELF+N F+DDTNVYRVT+HKTFEGN+ TK +NG IF NP+TG LFLKIIHT
Sbjct 1837 SSNYAELFNNDIKLFVDDTNVYRVTVHKTFEGNVATKAINGCIFTLNPKTGHLFLKIIHT 1896
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
SVWAGQKRL QLA+WKTAEEV+AL+RSLP EEQPKQII TRK MLDPLEVH+LDFPNI I
Sbjct 1897 SVWAGQKRLSQLAKWKTAEEVSALVRSLPKEEQPKQIIVTRKAMLDPLEVHMLDFPNIAI 1956
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
+ +EL LPF A M I+K D++++AT+P+MVLFN++DDWL IS+YTAFSRL LLLRAL+
Sbjct 1957 RPTELRLPFSAAMSIDKLSDVVMKATEPQMVLFNIYDDWLDRISSYTAFSRLTLLLRALK 2016
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
N E AK++L ++ + +H+WPS TDE+WI +E ++DLIL +Y +K NVN+++LTQ
Sbjct 2017 TNEESAKMILLSDPTITIKSYHLWPSFTDEQWITIESQMRDLILTEYGRKYNVNISALTQ 2076
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQ-------VTSTTTRSVNIHGEEMIVA 473
+EI+DIILG I PS++RQ++AE+EA E+Q T T+++N GEE++V
Sbjct 2077 TEIKDIILGQNIKAPSVKRQKMAELEAARSEKQNDEEAAGASTVMKTKTINAQGEEIVVV 2136
Query 474 TQSPHEQQVFSSKTDWRVRAISAASLHLRTRHLYVSSEAVAPAGCTYILPKNLLKKFISI 533
+ +E Q FSSK +WR AI+ L+LR +++YVS++ Y+LPKNLLKKFI I
Sbjct 2137 ASADYESQTFSSKNEWRKSAIANTLLYLRLKNIYVSADDFVEEQNVYVLPKNLLKKFIEI 2196
Query 534 ADLR 537
+D++
Sbjct 2197 SDVK 2200
> ECU04g0760
Length=2172
Score = 216 bits (549), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 142/457 (31%), Positives = 244/457 (53%), Gaps = 58/457 (12%)
Query 1 VHESIVMDLCQVFDMELDSLEVETVQKETIHPRKSYKMNSSCADILLFATYKWSISKPSL 60
+HES+V LC VF D V++ +H RKSY++ SSCADILL + + P
Sbjct 1546 IHESVVGGLCDVFRKYCD------VKRLEVHGRKSYRLRSSCADILLSGDF--CVDSPIS 1597
Query 61 LAEGKDIMEGATATKHWLDVQLRWGDYDSHDIERYCRSKFLDYTTDNMSIYPSPSGVLVG 120
+ E +D ++ W+DVQLRWGDYD + +Y R++F++ T D ++YP +G +V
Sbjct 1598 ILEERD-GGSVRCSELWIDVQLRWGDYDKRNPHKYARTRFVECTADPQALYPVKNGFVVV 1656
Query 121 VDLAYNLHSAFGNWIPGLKPLMQRAMNKIMKANPALYVLRERIRKGLQLYSSEPTEPYLN 180
+DL YN S +GN LK +++ +M +I+ + L++LRER+RK LQLY+S+ E N
Sbjct 1657 LDLCYNTWSGYGNLNEELKTVLKSSMERIVVEDAMLHILRERLRKALQLYTSD-IEVVSN 1715
Query 181 SQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHT 240
S G+LF++ + +D + R KT +F+ +P +G L+ K
Sbjct 1716 S---GDLFTSGLL--VDVKALLRK--EKT-------------LFVLDPASGNLYFK---- 1751
Query 241 SVWAGQKRLIQLARWKTAEEVAALIRSLPVEEQPKQIIATRKGMLDPLEVHLLDFPNIVI 300
++G+ + I+ + A++V L EE K+ I + M+D +E ++D P+I +
Sbjct 1752 -SYSGESKKIRQTKVLAAQDVFQL-----GEELNKRSIVVPESMIDAMENFIIDHPSISV 1805
Query 301 KGSELNLPFQALMRIEKFGDMILRATQPEMVLFNMFDDWLKSISAYTAFSRLLLLLRALQ 360
+ PF +M I++ +RA + + N+++ W ++ S +T F RLLL+++ +
Sbjct 1806 RTCLSAPPFSNVMGIKE-----IRAERSVRSI-NLYEGWGET-SRFTNFCRLLLVVQGMD 1858
Query 361 VNTERAKVLLRPHKSVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQ 420
V+ R + L + +WP +D EWI E+ LKDLI+ Y + + + L+Q
Sbjct 1859 VDEARVREL---------GINALWPGFSDSEWIKKEIQLKDLIVDRYCSMHGIEPSGLSQ 1909
Query 421 SEIRDIILGMEIAPPSLQRQQIAEIEAQAREQQQVTS 457
SE+RDI+ G ++ Q + I+ R +++ S
Sbjct 1910 SEVRDIVFGFRVSKG--QGATVETIDTGIRNSKKMLS 1944
> Hs10863953
Length=612
Score = 35.4 bits (80), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 34/150 (22%), Positives = 61/150 (40%), Gaps = 29/150 (19%)
Query 136 PGLKPLMQRAMNKIMKA-------NPALYVLRERIRKGLQLYSSEPTEPYLNSQNYGE-- 186
PG+ L + KA +P L E+I+ L +YS E + P++ ++Y E
Sbjct 235 PGIAALTSEERTRWAKAREYLIGLDPENLALLEKIQSSLLVYSMEDSSPHVTPEDYSEII 294
Query 187 ---LFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFNPRTGQLFLKIIHTSVW 243
L + TV + D + + F N P + +I +++ S +
Sbjct 295 AAILIGDPTVRWGDKSYNLISFSNGVFGCNCDHAPFDAMI-------------MVNISYY 341
Query 244 AGQKRLIQLARWKTAEEVAALIRSLPVEEQ 273
+K RWK +E+V R +P+ E+
Sbjct 342 VDEKIFQNEGRWKGSEKV----RDIPLPEE 367
> CE03783
Length=407
Score = 32.0 bits (71), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 50/108 (46%), Gaps = 15/108 (13%)
Query 324 RATQPEMVLFNMFDDWLKSISAYTAFSRLL--LLLRALQVN------TERAKVLLRPHKS 375
+ TQP+ + N D K + Y F++ + +LL A++ + + ++ L
Sbjct 70 KGTQPDKDVVNFVDKNYKPGTTYADFAKDVCNILLFAIESDFCSLPQSISMRINLLKLYF 129
Query 376 VVTQPHH----IWPSLTDEEWIHVEVGLKDLILG---DYAKKNNVNVA 416
V T HH +WPS T W +++G K I+G D KK +V+
Sbjct 130 VFTSKHHEGFTMWPSRTSWNWNSMDIGPKRDIVGELRDAFKKTDVHFG 177
> 7302060
Length=2090
Score = 32.0 bits (71), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 35/67 (52%), Gaps = 11/67 (16%)
Query 175 TEPYLNSQNYGELFSNQTVWFIDDTNVYRVTIHKTFEGNLTTKPVNGVIFIFN--PRTGQ 232
+PY+N N ELF+++ FI+D N+Y ++ V ++ F+ P+ +
Sbjct 1456 CQPYINEINNVELFTSKLKHFIEDINIYFYELYMC---------VINILMYFDSAPKEME 1506
Query 233 LFLKIIH 239
+++ I+H
Sbjct 1507 IWMNILH 1513
> YER140w
Length=556
Score = 31.2 bits (69), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Query 375 SVVTQPHHIWPSLTDEEWIHVEVGLKDLILGDYAKKNNVNVASLTQSEIRDIILGMEIAP 434
+VVT+ ++ P L V+V + + DY K+N + S++ R L E P
Sbjct 466 TVVTEEEYV-PGLLSGGMGKVDVSTRIALHSDYNKENRIETESVSPMRKRKTTLTAECTP 524
Query 435 PSL 437
PSL
Sbjct 525 PSL 527
Lambda K H
0.320 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14764723550
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40