bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1777_orf1
Length=189
Score E
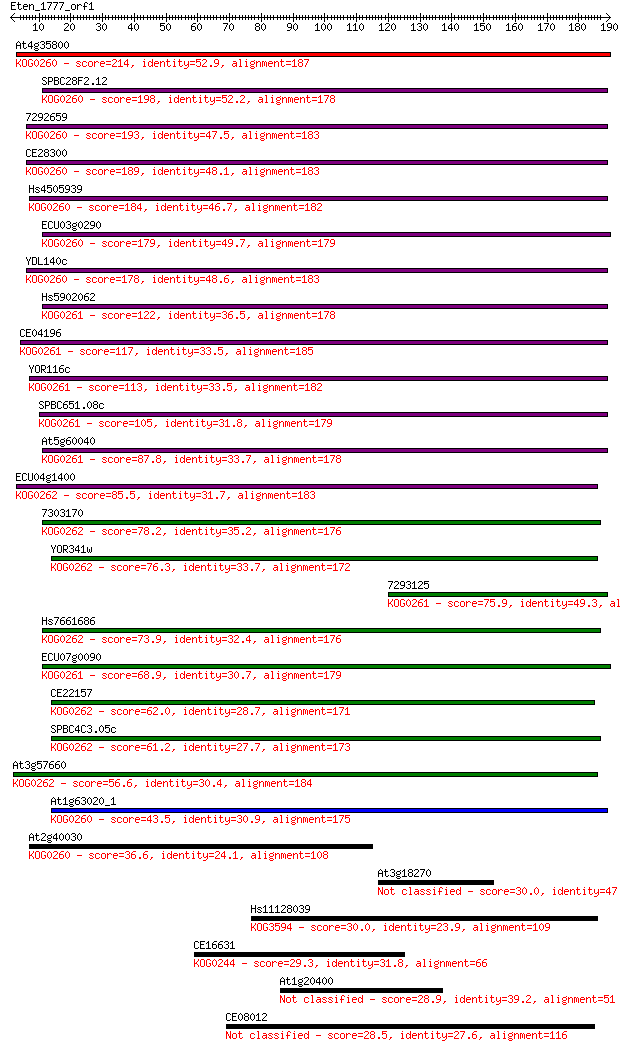
Sequences producing significant alignments: (Bits) Value
At4g35800 214 1e-55
SPBC28F2.12 198 6e-51
7292659 193 2e-49
CE28300 189 2e-48
Hs4505939 184 7e-47
ECU03g0290 179 4e-45
YDL140c 178 6e-45
Hs5902062 122 4e-28
CE04196 117 1e-26
YOR116c 113 2e-25
SPBC651.08c 105 7e-23
At5g60040 87.8 1e-17
ECU04g1400 85.5 6e-17
7303170 78.2 8e-15
YOR341w 76.3 3e-14
7293125 75.9 5e-14
Hs7661686 73.9 2e-13
ECU07g0090 68.9 6e-12
CE22157 62.0 8e-10
SPBC4C3.05c 61.2 1e-09
At3g57660 56.6 3e-08
At1g63020_1 43.5 2e-04
At2g40030 36.6 0.033
At3g18270 30.0 2.9
Hs11128039 30.0 3.0
CE16631 29.3 5.3
At1g20400 28.9 6.2
CE08012 28.5 8.4
> At4g35800
Length=1840
Score = 214 bits (544), Expect = 1e-55, Method: Composition-based stats.
Identities = 99/187 (52%), Positives = 132/187 (70%), Gaps = 0/187 (0%)
Query 3 ENLFVSERDERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLV 62
E F++ D +V I +GELLAG +CKKT+G+S+GSL+H++W E GP+ + FLG Q LV
Sbjct 614 ETGFITPGDTQVRIERGELLAGTLCKKTLGTSNGSLVHVIWEEVGPDAARKFLGHTQWLV 673
Query 63 NFWLLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRES 122
N+WLL GFT+G D I T +K+ E + AK V L R L+ +PG+++R++
Sbjct 674 NYWLLQNGFTIGIGDTIADSSTMEKINETISNAKTAVKDLIRQFQGKELDPEPGRTMRDT 733
Query 123 FEARVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGK 182
FE RVN+ LN AR+ +G A + L E+NN+ AMV AGSKGS INISQ+ ACVGQQNVEGK
Sbjct 734 FENRVNQVLNKARDDAGSSAQKSLAETNNLKAMVTAGSKGSFINISQMTACVGQQNVEGK 793
Query 183 RIPFGFN 189
RIPFGF+
Sbjct 794 RIPFGFD 800
> SPBC28F2.12
Length=1752
Score = 198 bits (503), Expect = 6e-51, Method: Composition-based stats.
Identities = 93/178 (52%), Positives = 125/178 (70%), Gaps = 0/178 (0%)
Query 11 DERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWLLHQG 70
D ++I GE++ G + KKTVG+S G L+H +W E GPE K F +Q++VN+WLLH G
Sbjct 608 DSGMLIENGEIIYGVVDKKTVGASQGGLVHTIWKEKGPEICKGFFNGIQRVVNYWLLHNG 667
Query 71 FTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVNKE 130
F++G D I +T K+V +KEA+ +VA + A RL+ +PG +LRESFEA+V++
Sbjct 668 FSIGIGDTIADADTMKEVTRTVKEARRQVAECIQDAQHNRLKPEPGMTLRESFEAKVSRI 727
Query 131 LNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIPFGF 188
LN AR+ +G+ A L +SNN+ MV AGSKGS INISQ+ ACVGQQ VEGKRIPFGF
Sbjct 728 LNQARDNAGRSAEHSLKDSNNVKQMVAAGSKGSFINISQMSACVGQQIVEGKRIPFGF 785
> 7292659
Length=1887
Score = 193 bits (490), Expect = 2e-49, Method: Composition-based stats.
Identities = 87/183 (47%), Positives = 133/183 (72%), Gaps = 0/183 (0%)
Query 6 FVSERDERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFW 65
++S D +V++ GEL+ G +CKK++G+S+GSL+H+ + E G + F G +Q ++N W
Sbjct 612 WISPGDTKVMVEHGELIMGILCKKSLGTSAGSLLHICFLELGHDIAGRFYGNIQTVINNW 671
Query 66 LLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEA 125
LL +G ++G D I +T ++++ +K+AKD+V ++ + AH LE PG +LR++FE
Sbjct 672 LLFEGHSIGIGDTIADPQTYNEIQQAIKKAKDDVINVIQKAHNMELEPTPGNTLRQTFEN 731
Query 126 RVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIP 185
+VN+ LN AR+K+G A + L E NN+ AMV++GSKGS INISQ++ACVGQQNVEGKRIP
Sbjct 732 KVNRILNDARDKTGGSAKKSLTEYNNLKAMVVSGSKGSNINISQVIACVGQQNVEGKRIP 791
Query 186 FGF 188
+GF
Sbjct 792 YGF 794
> CE28300
Length=1852
Score = 189 bits (481), Expect = 2e-48, Method: Composition-based stats.
Identities = 88/183 (48%), Positives = 126/183 (68%), Gaps = 0/183 (0%)
Query 6 FVSERDERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFW 65
++S D +VII GELL+G +C KTVG S+G+L+H++ E G E +F +Q ++N W
Sbjct 610 WISPGDTKVIIEHGELLSGIVCSKTVGKSAGNLLHVVTLELGYEIAANFYSHIQTVINAW 669
Query 66 LLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEA 125
L+ +G T+G D I Q T ++ +++AK +V + AH LE PG +LR++FE
Sbjct 670 LIREGHTIGIGDTIADQATYLDIQNTIRKAKQDVVDVIEKAHNDDLEPTPGNTLRQTFEN 729
Query 126 RVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIP 185
+VN+ LN AR+++G A + L E NN +MV++GSKGS INISQ++ACVGQQNVEGKRIP
Sbjct 730 KVNQILNDARDRTGSSAQKSLSEFNNFKSMVVSGSKGSKINISQVIACVGQQNVEGKRIP 789
Query 186 FGF 188
FGF
Sbjct 790 FGF 792
> Hs4505939
Length=1970
Score = 184 bits (468), Expect = 7e-47, Method: Composition-based stats.
Identities = 85/182 (46%), Positives = 127/182 (69%), Gaps = 0/182 (0%)
Query 7 VSERDERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWL 66
+S D +V++ GEL+ G +CKK++G+S+GSL+H+ + E G + T+ F +Q ++N WL
Sbjct 621 ISPGDTKVVVENGELIMGILCKKSLGTSAGSLVHISYLEMGHDITRLFYSNIQTVINNWL 680
Query 67 LHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEAR 126
L +G T+G D I +T + ++ +K+AK +V + AH LE PG +LR++FE +
Sbjct 681 LIEGHTIGIGDSIADSKTYQDIQNTIKKAKQDVIEVIEKAHNNELEPTPGNTLRQTFENQ 740
Query 127 VNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIPF 186
VN+ LN AR+K+G A + L E NN +MV++G+KGS INISQ++A VGQQNVEGKRIPF
Sbjct 741 VNRILNDARDKTGSSAQKSLSEYNNFKSMVVSGAKGSKINISQVIAVVGQQNVEGKRIPF 800
Query 187 GF 188
GF
Sbjct 801 GF 802
> ECU03g0290
Length=1599
Score = 179 bits (453), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 89/181 (49%), Positives = 125/181 (69%), Gaps = 3/181 (1%)
Query 11 DERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWL--LH 68
D RVII+ G + +G I KK G++ G L+H+++N+ GP+R F +Q+++N ++ +H
Sbjct 595 DTRVIIQDGYIHSGVIDKKAAGATQGGLVHIIFNDFGPKRAAQFFDGVQRMINAFMTGIH 654
Query 69 QGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVN 128
F++G D I +T K VE +++AK+EV+ L A Q RLE PG +++ESFE+ +N
Sbjct 655 T-FSMGIGDTIADPKTVKVVESAIRKAKEEVSALIENARQNRLERLPGMTMKESFESHLN 713
Query 129 KELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIPFGF 188
LN AR+ SG A L E+NN+ MVLAGSKGS INISQ+ ACVGQQNVEGKRIPFGF
Sbjct 714 LVLNRARDVSGTSAQRSLSENNNMKTMVLAGSKGSFINISQVTACVGQQNVEGKRIPFGF 773
Query 189 N 189
+
Sbjct 774 S 774
> YDL140c
Length=1733
Score = 178 bits (451), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 89/183 (48%), Positives = 124/183 (67%), Gaps = 0/183 (0%)
Query 6 FVSERDERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFW 65
+S +D ++I G+++ G + KKTVGSS+G LIH++ E GP+ G +QK+VNFW
Sbjct 597 LLSPKDNGMLIIDGQIIFGVVEKKTVGSSNGGLIHVVTREKGPQVCAKLFGNIQKVVNFW 656
Query 66 LLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEA 125
LLH GF+ G D I T +++ E + EAK +V +T+ A L A+ G +LRESFE
Sbjct 657 LLHNGFSTGIGDTIADGPTMREITETIAEAKKKVLDVTKEAQANLLTAKHGMTLRESFED 716
Query 126 RVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIP 185
V + LN AR+K+G++A L + NN+ MV+AGSKGS INI+Q+ ACVGQQ+VEGKRI
Sbjct 717 NVVRFLNEARDKAGRLAEVNLKDLNNVKQMVMAGSKGSFINIAQMSACVGQQSVEGKRIA 776
Query 186 FGF 188
FGF
Sbjct 777 FGF 779
> Hs5902062
Length=1391
Score = 122 bits (307), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 65/179 (36%), Positives = 102/179 (56%), Gaps = 1/179 (0%)
Query 11 DERVIIRQGELLAGKICKKTVGS-SSGSLIHLLWNEAGPERTKDFLGALQKLVNFWLLHQ 69
D V I+ EL++G + K T+GS S ++ ++L + G D + L +L +L ++
Sbjct 635 DSYVTIQNSELMSGSMDKGTLGSGSKNNIFYILLRDWGQNEAADAMSRLARLAPVYLSNR 694
Query 70 GFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVNK 129
GF++G D+ Q K E+L + + G+L+ QPG + E+ EA + K
Sbjct 695 GFSIGIGDVTPGQGLLKAKYELLNAGYKKCDEYIEALNTGKLQQQPGCTAEETLEALILK 754
Query 130 ELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIPFGF 188
EL+ R+ +G +LD+SN+ + M L GSKGS INISQ++ACVGQQ + G R+P GF
Sbjct 755 ELSVIRDHAGSACLRELDKSNSPLTMALCGSKGSFINISQMIACVGQQAISGSRVPDGF 813
> CE04196
Length=1388
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 62/186 (33%), Positives = 110/186 (59%), Gaps = 1/186 (0%)
Query 4 NLFVSERDERVIIRQGELLAGKICKKTVGSSSG-SLIHLLWNEAGPERTKDFLGALQKLV 62
NL + +D VIIR LLAG + K +GSSS ++ ++L + G + D + L ++
Sbjct 624 NLELCSKDSYVIIRNSVLLAGCLDKSLLGSSSKVNIFYMLMRDYGEDAAVDAMWRLARMA 683
Query 63 NFWLLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRES 122
+L ++GF++G D+ S++ ++ +++ + + + +G+L+AQPG + E+
Sbjct 684 PVFLSNRGFSIGIGDVRPSEQLLREKGQLVDNGYSKCSQYIKELEEGKLKAQPGCTEEET 743
Query 123 FEARVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGK 182
E+ + +EL+ R+ +G++ L + N + M + GSKGS INISQ++ACVGQQ + G
Sbjct 744 LESIILRELSTIRDHAGQVCLRNLSKYNAPLTMAVCGSKGSFINISQMIACVGQQAISGH 803
Query 183 RIPFGF 188
R P GF
Sbjct 804 RPPDGF 809
> YOR116c
Length=1460
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 61/183 (33%), Positives = 108/183 (59%), Gaps = 1/183 (0%)
Query 7 VSERDERVIIRQGELLAGKICKKTVGSSS-GSLIHLLWNEAGPERTKDFLGALQKLVNFW 65
+S+ D VIIR ++L+G + K +G S+ + + + GP+ + + + KL +
Sbjct 645 MSQNDGFVIIRGSQILSGVMDKSVLGDGKKHSVFYTILRDYGPQEAANAMNRMAKLCARF 704
Query 66 LLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEA 125
L ++GF++G +D+ + + +K EE+++ A + L L ++G LE QPG + ++ EA
Sbjct 705 LGNRGFSIGINDVTPADDLKQKKEELVEIAYHKCDELITLFNKGELETQPGCNEEQTLEA 764
Query 126 RVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIP 185
++ L+ RE+ G + +LD N + M GSKGST+N+SQ++A VGQQ + G R+P
Sbjct 765 KIGGLLSKVREEVGDVCINELDNWNAPLIMATCGSKGSTLNVSQMVAVVGQQIISGNRVP 824
Query 186 FGF 188
GF
Sbjct 825 DGF 827
> SPBC651.08c
Length=1405
Score = 105 bits (261), Expect = 7e-23, Method: Composition-based stats.
Identities = 57/180 (31%), Positives = 99/180 (55%), Gaps = 1/180 (0%)
Query 10 RDERVIIRQGELLAGKICKKTVGS-SSGSLIHLLWNEAGPERTKDFLGALQKLVNFWLLH 68
+D ++IR E++AG + K VG SL +++ + G + + L K+ +L +
Sbjct 628 KDGYLMIRNSEIIAGVVDKSVVGDGKKDSLFYVILRDYGALEAAEAITRLSKMCARFLGN 687
Query 69 QGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVN 128
+GF++G D+ + + E ++ +A +G LE QPG + EA+++
Sbjct 688 RGFSIGIEDVQPGKSLSSQKEILVNKAYATSDDFIMQYAKGILECQPGMDQEATLEAKIS 747
Query 129 KELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIPFGF 188
L+ R+ G+I ++L +N+ + M GSKGS IN+SQ++ACVGQQ + GKR+P GF
Sbjct 748 STLSKVRDDVGEICMDELGPANSPLIMATCGSKGSKINVSQMVACVGQQIISGKRVPDGF 807
> At5g60040
Length=1328
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 60/187 (32%), Positives = 91/187 (48%), Gaps = 10/187 (5%)
Query 11 DERVIIRQGELLAGKICKKTV---------GSSSGSLIHLLWNEAGPERTKDFLGALQKL 61
D V R EL++G++ K T+ G+ G L +L + + L KL
Sbjct 605 DGWVYFRNSELISGQLGKATLALDIFPLGNGNKDG-LYSILLRDYNSHAAAVCMNRLAKL 663
Query 62 VNFWLLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRE 121
W+ GF++G D+ +E K+ ++ ++ D+ ++G L+ + G +
Sbjct 664 SARWIGIHGFSIGIDDVQPGEELSKERKDSIQFGYDQCHRKIEEFNRGNLQLKAGLDGAK 723
Query 122 SFEARVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEG 181
S EA + LN RE +GK L N+ + M GSKGS INISQ++ACVGQQ V G
Sbjct 724 SLEAEITGILNTIREATGKACMSGLHWRNSPLIMSQCGSKGSPINISQMVACVGQQTVNG 783
Query 182 KRIPFGF 188
R P GF
Sbjct 784 HRAPDGF 790
> ECU04g1400
Length=1395
Score = 85.5 bits (210), Expect = 6e-17, Method: Composition-based stats.
Identities = 58/192 (30%), Positives = 101/192 (52%), Gaps = 13/192 (6%)
Query 3 ENLFVSERDERVI-IRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKL 61
+N + +ER++ IR+G ++ G + K ++G + SLIH G + D L + +
Sbjct 609 KNAWREHSEERILRIREGNIVTGILDKNSLGPTFKSLIHACGEIKGFATSNDLLTYIGWV 668
Query 62 VNFWLLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSL-- 119
N +LL GFT+G D++ +E KK E+++ E L R +EA P L
Sbjct 669 GNRYLLMYGFTIGIDDLLLDKEADKKRREVVRVKDQEAKELQRR----YVEANPDFYLYA 724
Query 120 --RESFEARVNKELNAAREKSGKIA-SEQLDES---NNIMAMVLAGSKGSTINISQIMAC 173
+ ++ + E+N+ + K++ L +S NN+ ++ GSKGS +N+SQI
Sbjct 725 DKKAYLDSVMRTEMNSVTSEIVKVSVPSGLQKSFPENNMELIIATGSKGSIVNLSQISGA 784
Query 174 VGQQNVEGKRIP 185
+GQQ +EG+R+P
Sbjct 785 LGQQELEGQRVP 796
> 7303170
Length=1642
Score = 78.2 bits (191), Expect = 8e-15, Method: Composition-based stats.
Identities = 62/194 (31%), Positives = 95/194 (48%), Gaps = 19/194 (9%)
Query 11 DERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWLLHQG 70
+ +V IR GELL G + K+ G+++ LIH ++ G + + L A K+ F+L +G
Sbjct 714 ESQVQIRNGELLVGVLDKQQYGATTYGLIHCMYELYGGDVSTLLLTAFTKVFTFFLQLEG 773
Query 71 FTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVNKE 130
FT+G DI+ + +K +I++E ++ V + A + P L E EA K+
Sbjct 774 FTLGVKDILVTDVADRKRRKIIRECRN-VGNSAVAAALELEDEPPHDELVEKMEAAYVKD 832
Query 131 -----LNAAREKS------GKIASEQLDE-------SNNIMAMVLAGSKGSTINISQIMA 172
L + KS I S L SNN+ MVL+G+KGS +N QI
Sbjct 833 SKFRVLLDRKYKSLLDGYTNDINSTCLPRGLITKFPSNNLQLMVLSGAKGSMVNTMQISC 892
Query 173 CVGQQNVEGKRIPF 186
+GQ +EGKR P
Sbjct 893 LLGQIELEGKRPPL 906
> YOR341w
Length=1664
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 58/193 (30%), Positives = 96/193 (49%), Gaps = 24/193 (12%)
Query 14 VIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWLLHQGFTV 73
V+ + G LL G + K G+S ++H L GPE L L +L ++ FT
Sbjct 769 VLFKDGALLCGILDKSQYGASKYGIVHSLHEVYGPEVAAKVLSVLGRLFTNYITATAFTC 828
Query 74 GCSDIITSQETCKKVEEILK-------EAKDEVAHLTR--LAHQGRLEAQPGKSLRES-- 122
G D+ + E K +ILK EA EV +L + A L + + LR++
Sbjct 829 GMDDLRLTAEGNKWRTDILKTSVDTGREAAAEVTNLDKDTPADDPELLKRLQEILRDNNK 888
Query 123 ---FEARVNKELNAAREKSGKIASEQLDES-------NNIMAMVLAGSKGSTINISQIMA 172
+A + ++NA + ++ S+ + + N++ AM L+G+KGS +N+SQIM
Sbjct 889 SGILDAVTSSKVNAI---TSQVVSKCVPDGTMKKFPCNSMQAMALSGAKGSNVNVSQIMC 945
Query 173 CVGQQNVEGKRIP 185
+GQQ +EG+R+P
Sbjct 946 LLGQQALEGRRVP 958
> 7293125
Length=1259
Score = 75.9 bits (185), Expect = 5e-14, Method: Composition-based stats.
Identities = 34/69 (49%), Positives = 51/69 (73%), Gaps = 0/69 (0%)
Query 120 RESFEARVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNV 179
+E+ E+ + +EL+A RE++ K +L +N+ + M L+GSKGS INISQ++ACVGQQ +
Sbjct 619 KETLESVMLRELSAIREQAAKTCFAELHPTNSALIMALSGSKGSNINISQMIACVGQQAI 678
Query 180 EGKRIPFGF 188
GKR+P GF
Sbjct 679 SGKRVPNGF 687
> Hs7661686
Length=1717
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 57/196 (29%), Positives = 91/196 (46%), Gaps = 21/196 (10%)
Query 11 DERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWL-LHQ 69
+ +VIIR+GELL G + K GSS+ L+H + G E + L L +L +L L++
Sbjct 734 ESQVIIREGELLCGVLDKAHYGSSAYGLVHCCYEIYGGETSGKVLTCLARLFTAYLQLYR 793
Query 70 GFTVGCSDIITSQETCKKVEEILKEAK-------------------DEVAHLTRLAHQGR 110
GFT+G DI+ + K + I++E+ DEV + AH G+
Sbjct 794 GFTLGVEDILVKPKRDVKRQRIIEESTHCGPQAVRAALNLPEAASYDEVRGKWQDAHLGK 853
Query 111 LEAQPGKSLRESFEARVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQI 170
+ + + F+ VN N + + N + MV +G+KGST+N QI
Sbjct 854 -DQRDFNMIDLKFKEEVNHYSNEINKACMPFGLHRQFPENTLQLMVQSGAKGSTVNTMQI 912
Query 171 MACVGQQNVEGKRIPF 186
+GQ +EG+ P
Sbjct 913 SCLLGQIELEGRSTPL 928
> ECU07g0090
Length=1349
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 55/181 (30%), Positives = 86/181 (47%), Gaps = 9/181 (4%)
Query 11 DERVIIRQGELLAGKICKKTVGSSS--GSLIHLLWNEAGPERTKDFLGALQKLVNFWLLH 68
D V I G++ K VG + SLI+ + + + + ++ KL + +L
Sbjct 639 DGSVAILDNSYYFGRLDKSIVGGENKRDSLIYAIMKVSSMAAVRA-MNSITKLCSRYLGE 697
Query 69 QGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVN 128
GF++G D+ +K E +++ E L + + EA E E ++
Sbjct 698 TGFSIGLDDVQPGPILRQKKEMVVRRGYAECESKI-LEYSKKPEAN-----EEMLEMEIS 751
Query 129 KELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIPFGF 188
LN RE+ G I ++L N+ M GSKGS IN+SQ++ACVGQQ V G RIP G
Sbjct 752 SILNRIREECGSICIKELGIRNSPNIMQACGSKGSKINVSQMVACVGQQIVSGTRIPNGM 811
Query 189 N 189
+
Sbjct 812 D 812
> CE22157
Length=1737
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 49/191 (25%), Positives = 80/191 (41%), Gaps = 20/191 (10%)
Query 14 VIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWLLHQGFTV 73
V+ RQGELL G + K G++ L H + G L +L +L GFT+
Sbjct 772 VVFRQGELLVGVLDKAHFGATQFGLAHCAFELYGHRCGVQLLSCFSRLFTTYLQFHGFTL 831
Query 74 GCSDIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPG----------------- 116
G +DI+ ++ K +E + E++ + + A A P
Sbjct 832 GVADILVVKDADGKRKEAVMESRTIGNQVVKTAFGLPDTATPAEIKRTLAATYCNPRGQG 891
Query 117 ---KSLRESFEARVNKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMAC 173
K L + + K +A + +L N + M+ +G+KGS +N QI C
Sbjct 892 TDVKMLDFGMKQGIAKYNDAITKSCVPTGLLRLFPQNALQLMIQSGAKGSAVNAIQISGC 951
Query 174 VGQQNVEGKRI 184
+GQ +EGKR+
Sbjct 952 LGQIELEGKRM 962
> SPBC4C3.05c
Length=1689
Score = 61.2 bits (147), Expect = 1e-09, Method: Composition-based stats.
Identities = 48/191 (25%), Positives = 88/191 (46%), Gaps = 20/191 (10%)
Query 14 VIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWLLHQGFTV 73
V+ GELL G + K + G+S+ L+H + GP+ L L +L + +GFT
Sbjct 784 VLFDDGELLCGILDKSSFGASAFGLVHSVHELYGPDIAGRLLSVLSRLFTAYAQMRGFTC 843
Query 74 GCSDIITSQETCKKVEEILK-------EAKDEVAHLTRLAHQGRLEAQPGKSLRESFEAR 126
D+ ++ ++L+ EA E L+ + L A + R+ + +
Sbjct 844 RMDDLRLDEQGDNWRRQLLENGKSFGLEAASEYVGLSTDSPIALLNANLEEVYRD--DEK 901
Query 127 VNKELNAAREKSGKIASEQLDES-----------NNIMAMVLAGSKGSTINISQIMACVG 175
+ A + K + S +++ N++ M ++G+KGS +N+SQI +G
Sbjct 902 LQGLDAAMKGKMNGLTSSIINKCIPDGLLTKFPYNHMQTMTVSGAKGSNVNVSQISCLLG 961
Query 176 QQNVEGKRIPF 186
QQ +EG+R+P
Sbjct 962 QQELEGRRVPL 972
> At3g57660
Length=1670
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 56/205 (27%), Positives = 94/205 (45%), Gaps = 27/205 (13%)
Query 2 KENLFVSERDERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKL 61
K+NL ++++ IR+ E + G I K L+H + G + L +L
Sbjct 773 KQNL----NEDKLHIRKNEFVCGVIDKAQFADYG--LVHTVHELYGSNAAGNLLSVFSRL 826
Query 62 VNFWLLHQGFTVGCSDII---------TSQ-ETCKKV-EEILKEA--KDEVAHLTRLAHQ 108
+L GFT G D+I T Q + C+ V E +L++ D + +
Sbjct 827 FTVFLQTHGFTCGVDDLIILKDMDEERTKQLQECENVGERVLRKTFGIDVDVQIDPQDMR 886
Query 109 GRLE---AQPGKSLRESFEARVNKELNAAREKS--GKIASEQLDES---NNIMAMVLAGS 160
R+E + G+S S + + LN K + S+ L ++ N I M ++G+
Sbjct 887 SRIERILYEDGESALASLDRSIVNYLNQCSSKGVMNDLLSDGLLKTPGRNCISLMTISGA 946
Query 161 KGSTINISQIMACVGQQNVEGKRIP 185
KGS +N QI + +GQQ++EGKR+P
Sbjct 947 KGSKVNFQQISSHLGQQDLEGKRVP 971
> At1g63020_1
Length=1316
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 54/208 (25%), Positives = 90/208 (43%), Gaps = 44/208 (21%)
Query 14 VIIRQGELLAGKICKKTVGSSSGSLIH-LLWNEAGPERTKDFLGALQKLVNFWLLHQGFT 72
V++ GELL+ + G+ I LL ++ G + D + + Q++++ WLL +G +
Sbjct 564 VVVSNGELLSFSEGSAWLRDGEGNFIERLLKHDKG--KVLDIIYSAQEMLSQWLLMRGLS 621
Query 73 VGCSDIITSQETCKK---VEEI---LKEAK--------------------------DEVA 100
V +D+ S + + EEI L+EA+ D V+
Sbjct 622 VSLADLYLSSDLQSRKNLTEEISYGLREAEQVCNKQQLMVESWRDFLAVNGEDKEEDSVS 681
Query 101 HLTRLAHQGRLEAQPGKSLRESFEARVNKELNAAREKSGKIASEQLDESNNIMAMVLAGS 160
L R + E Q +L E V+ +A R+ +A D+SN+ + M AGS
Sbjct 682 DLARFCY----ERQKSATLSE---LAVSAFKDAYRDVQA-LAYRYGDQSNSFLIMSKAGS 733
Query 161 KGSTINISQIMACVGQQNVEGKRIPFGF 188
KG+ + Q C+G QN + FGF
Sbjct 734 KGNIGKLVQHSMCIGLQN-SAVSLSFGF 760
> At2g40030
Length=888
Score = 36.6 bits (83), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 54/109 (49%), Gaps = 2/109 (1%)
Query 7 VSERDERVIIRQGELLAGKICKKTVGSSSGSLIHLLWNEAGPERTKDFLGALQKLVNFWL 66
+S + +R ++ +LL +GS ++ ++ E GP+ T F +LQ L+ L
Sbjct 504 LSCKGDRFLVDGSDLLKFDFGVDAMGSIINEIVTSIFLEKGPKETLGFFDSLQPLLMESL 563
Query 67 LHQGFTVGCSDIITSQETCKKVEE-ILKEAKDEVAHLTRLAHQGRLEAQ 114
+GF++ D+ S+ + I++E V+ L RL+++ L+ +
Sbjct 564 FAEGFSLSLEDLSMSRADMDVIHNLIIREISPMVSRL-RLSYRDELQLE 611
> At3g18270
Length=410
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 23/40 (57%), Gaps = 4/40 (10%)
Query 117 KSLRESFEARV----NKELNAAREKSGKIASEQLDESNNI 152
K+L E+F RV N+ELN A IAS +LD +N+
Sbjct 38 KTLTENFTVRVLKAENRELNVALLSPFTIASSRLDSVSNV 77
> Hs11128039
Length=935
Score = 30.0 bits (66), Expect = 3.0, Method: Composition-based stats.
Identities = 26/109 (23%), Positives = 45/109 (41%), Gaps = 2/109 (1%)
Query 77 DIITSQETCKKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVNKELNAARE 136
D + SQE+C+K E +L A+D L + +A P R S R + +
Sbjct 778 DTLISQESCEKSEPLLI-AEDSAIILGKCDPTSNQQAPPNTDWRFSQAQRPGTSGSQNGD 836
Query 137 KSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRIP 185
+G + Q D + + AM+LA + + S + G + + P
Sbjct 837 DTGTWPNNQFD-TEMLQAMILASASEAADGSSTLGGGAGTMGLSARYGP 884
> CE16631
Length=1083
Score = 29.3 bits (64), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 36/70 (51%), Gaps = 4/70 (5%)
Query 59 QKLVNFWLLHQGFTVGCSDIITSQETCKKVEEILKEAKDEVAHLT----RLAHQGRLEAQ 114
QKL + H +D+ QET +++EE LK+ +DE+ +L RL + R EA
Sbjct 565 QKLKDLERQHAEDKKVLNDMKKLQETRRRMEETLKKTEDELKNLKTQRLRLLREQRAEAS 624
Query 115 PGKSLRESFE 124
++ ++ E
Sbjct 625 KFQAFKQKHE 634
> At1g20400
Length=944
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 25/51 (49%), Gaps = 5/51 (9%)
Query 86 KKVEEILKEAKDEVAHLTRLAHQGRLEAQPGKSLRESFEARVNKELNAARE 136
+ VEE + A+ E HQGR PG S+ S E R KEL AR+
Sbjct 554 RSVEEERRTARSEPPR----GHQGRSVVNPGSSVDRSKETR-EKELATARQ 599
> CE08012
Length=384
Score = 28.5 bits (62), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 52/117 (44%), Gaps = 6/117 (5%)
Query 69 QGFTVGCSDIITSQETCKKVEEILKEAKDEVA-HLTRLAHQGRLEAQPGKSLRESFEARV 127
QG TV SD S ++ KK + E E+ +L + R+ K+ R + A
Sbjct 146 QGCTVSYSDNRASSDSRKKEINVTDEDYVELCIKDLKLLMENRMVHL--KTFRFAIIAVA 203
Query 128 NKELNAAREKSGKIASEQLDESNNIMAMVLAGSKGSTINISQIMACVGQQNVEGKRI 184
KE REK K E L + ++ + + + G + ++Q+MA +NVE R
Sbjct 204 TKEF---REKCMKRVEEALKTAKSLTSRTVETTGGYLLELAQLMAIFPAKNVEEIRF 257
Lambda K H
0.315 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3130490202
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40