bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1805_orf5
Length=232
Score E
Sequences producing significant alignments: (Bits) Value
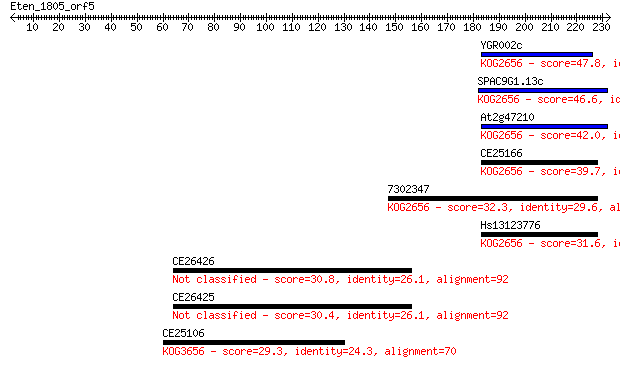
YGR002c 47.8 2e-05
SPAC9G1.13c 46.6 4e-05
At2g47210 42.0 0.001
CE25166 39.7 0.005
7302347 32.3 0.75
Hs13123776 31.6 1.3
CE26426 30.8 2.7
CE26425 30.4 2.8
CE25106 29.3 6.7
> YGR002c
Length=476
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 30/43 (69%), Gaps = 0/43 (0%)
Query 183 WSFEATLELWQLCEVYELRWFVIADRFKENNKMKITDLKKRYF 225
WSFE L+ LC+ Y+LRWF+I DR+ NN + DLK++++
Sbjct 164 WSFEEIEYLFNLCKKYDLRWFLIFDRYSYNNSRTLEDLKEKFY 206
> SPAC9G1.13c
Length=411
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 39/54 (72%), Gaps = 5/54 (9%)
Query 182 EWSFEATLELWQLCEVYELRWFVIADRFKENNKMK----ITDLKKRYFAVAKVL 231
+W+ + T L++LC+ Y+LR+FVIADR+ +N K K + DLK R+++V++ +
Sbjct 101 DWNKDETDYLFRLCKDYDLRFFVIADRY-DNEKYKKHRTLEDLKDRFYSVSRKI 153
> At2g47210
Length=486
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 33/49 (67%), Gaps = 2/49 (4%)
Query 183 WSFEATLELWQLCEVYELRWFVIADRFKENNKMKITDLKKRYFAVAKVL 231
W+ E T +L++ C+ ++LR+ VIADRF + + +LK RY++V + L
Sbjct 130 WTKEETDQLFEFCQNFDLRFVVIADRFPVS--RTVEELKDRYYSVNRAL 176
> CE25166
Length=562
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 30/48 (62%), Gaps = 3/48 (6%)
Query 183 WSFEATLELWQLCEVYELRWFVIADRF---KENNKMKITDLKKRYFAV 227
WS E T L+ C +++LRW ++ DRF K N + DLK+R++++
Sbjct 134 WSREETDYLFDTCRMFDLRWPIVYDRFDCKKFNQNRTVEDLKERFYSI 181
> 7302347
Length=433
Score = 32.3 bits (72), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 45/83 (54%), Gaps = 2/83 (2%)
Query 147 VDEDAEFNYAKFHVKIRHPPLTKALYDKLLADLDPEWSFEATLELWQLCEVYELRWFVIA 206
D ++ +AKF+ ++ P T Y+ L + WS T L+ L ++LR+ V+A
Sbjct 114 TDNSTDYPFAKFNKQLEVPSYTMTEYNAHLRNNINNWSKVQTDHLFDLARRFDLRFIVMA 173
Query 207 DRF--KENNKMKITDLKKRYFAV 227
DR+ +++ + +LK+RY+ V
Sbjct 174 DRWNRQQHGTKTVEELKERYYEV 196
> Hs13123776
Length=467
Score = 31.6 bits (70), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Query 183 WSFEATLELWQLCEVYELRWFVIADRFKENN--KMKITDLKKRYFAV 227
W+ T L+ L ++LR+ VI DR+ K + DLK+RY+ +
Sbjct 152 WTKAETDHLFDLSRRFDLRFVVIHDRYDHQQFKKRSVEDLKERYYHI 198
> CE26426
Length=351
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 42/96 (43%), Gaps = 4/96 (4%)
Query 64 VLLVSLLVLLLVLLLVVLLLLLLLVVLLLLVVLPLLRVPILVLLLVLLLMLLLVVLLLLL 123
V ++ V+ +V V +++VL LVV PLL PI V +LM ++ L++
Sbjct 241 VYFSTMFVITVVFYHVAFFCSYIILVLTFLVVGPLLFNPIDVWAGNGILMTIMATYNLVI 300
Query 124 QL----LLQLLLLLLQMELNDRLKYFQVDEDAEFNY 155
+ L + + ELN +++ NY
Sbjct 301 GIVAVNLYNKIQTIKSEELNGKVQIQTTGNTGAQNY 336
> CE26425
Length=344
Score = 30.4 bits (67), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 42/96 (43%), Gaps = 4/96 (4%)
Query 64 VLLVSLLVLLLVLLLVVLLLLLLLVVLLLLVVLPLLRVPILVLLLVLLLMLLLVVLLLLL 123
V ++ V+ +V V +++VL LVV PLL PI V +LM ++ L++
Sbjct 234 VYFSTMFVITVVFYHVAFFCSYIILVLTFLVVGPLLFNPIDVWAGNGILMTIMATYNLVI 293
Query 124 QL----LLQLLLLLLQMELNDRLKYFQVDEDAEFNY 155
+ L + + ELN +++ NY
Sbjct 294 GIVAVNLYNKIQTIKSEELNGKVQIQTTGNTGAQNY 329
> CE25106
Length=422
Score = 29.3 bits (64), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 60 SLLVVLLVSLLVLLLVLLLVVLLLLLLLVVLLLLVVLPLLRVPILVLLLVLLLMLLLVVL 119
S + V + V+++ L+ V+ L + L+++ P++R P L L + LLV +
Sbjct 26 SFIDQPFVKIPVMIIYCLVFVVCLSGNFLTLIVMTSHPMMRTPTNFFLSNLAIADLLVAI 85
Query 120 LLLLQLLLQL 129
+LQ ++ +
Sbjct 86 FCILQNMIHI 95
Lambda K H
0.337 0.154 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4526773522
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40