bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1813_orf3
Length=194
Score E
Sequences producing significant alignments: (Bits) Value
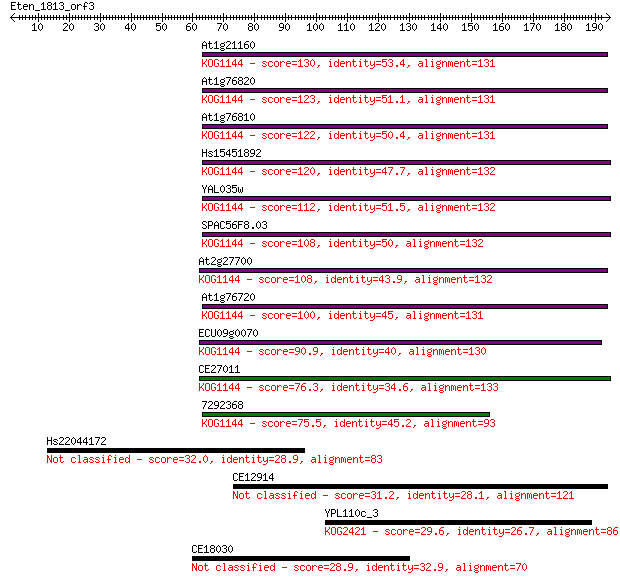
At1g21160 130 1e-30
At1g76820 123 2e-28
At1g76810 122 4e-28
Hs15451892 120 2e-27
YAL035w 112 4e-25
SPAC56F8.03 108 7e-24
At2g27700 108 9e-24
At1g76720 100 1e-21
ECU09g0070 90.9 1e-18
CE27011 76.3 4e-14
7292368 75.5 6e-14
Hs22044172 32.0 0.80
CE12914 31.2 1.4
YPL110c_3 29.6 3.9
CE18030 28.9 7.1
> At1g21160
Length=1088
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 70/133 (52%), Positives = 90/133 (67%), Gaps = 3/133 (2%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
EMRV G Y H V+AA G+KI A GLE A+AGT+L V ED+ E + M+++ V+
Sbjct 772 EMRVTGTYMPHREVKAAQGIKIAAQGLEHAIAGTALHVIGPNEDMEEAKKNAMEDIESVM 831
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLE--DCKIPVFAVNIGTVQKKDIKKASVMREKGF 180
+++K+ GVYV ASTLG+LEALL +L+ D KIPV + IG V KKDI KA VM EK
Sbjct 832 NRIDKSGEGVYVQASTLGSLEALLEFLKSSDVKIPVSGIGIGPVHKKDIMKAGVMLEKK- 890
Query 181 AEFAVVLGFDVKV 193
EFA +L FDVK+
Sbjct 891 KEFATILAFDVKI 903
> At1g76820
Length=1146
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 67/133 (50%), Positives = 89/133 (66%), Gaps = 3/133 (2%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG Y H ++AA G+KI A GLE A+AGTSL V ++DI ++ M+++ VL
Sbjct 853 ELRVKGTYLHHKEIKAAQGIKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVL 912
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLED--CKIPVFAVNIGTVQKKDIKKASVMREKGF 180
+++K+ GVYV STLG+LEALL +L+ IPV + IG V KKDI KA VM EK
Sbjct 913 SRIDKSGEGVYVQTSTLGSLEALLEFLKTPAVNIPVSGIGIGPVHKKDIMKAGVMLEKK- 971
Query 181 AEFAVVLGFDVKV 193
E+A +L FDVKV
Sbjct 972 KEYATILAFDVKV 984
> At1g76810
Length=1280
Score = 122 bits (306), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 66/133 (49%), Positives = 91/133 (68%), Gaps = 3/133 (2%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG Y + ++AA G+KI A GLE A+AGT+L V ++DI ++ M+++ VL
Sbjct 970 ELRVKGTYLHYKEIKAAQGIKITAQGLEHAIAGTALHVVGPDDDIEAIKESAMEDMESVL 1029
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLED--CKIPVFAVNIGTVQKKDIKKASVMREKGF 180
+++K+ GVYV ASTLG+LEALL YL+ KIPV + IG V KKD+ KA VM E+
Sbjct 1030 SRIDKSGEGVYVQASTLGSLEALLEYLKSPAVKIPVSGIGIGPVHKKDVMKAGVMLERK- 1088
Query 181 AEFAVVLGFDVKV 193
E+A +L FDVKV
Sbjct 1089 KEYATILAFDVKV 1101
> Hs15451892
Length=1220
Score = 120 bits (301), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 63/132 (47%), Positives = 88/132 (66%), Gaps = 1/132 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVK Y+ H VEAA GVKI+ LE +AG L VA KE++I L+ E++ E+ L
Sbjct 912 ELRVKNQYEKHKEVEAAQGVKILGKDLEKTLAGLPLLVAYKEDEIPVLKDELIHELKQTL 971
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
++ GVYV ASTLG+LEALL +L+ ++P +NIG V KKD+ KASVM E +
Sbjct 972 NAIKLEEKGVYVQASTLGSLEALLEFLKTSEVPYAGINIGPVHKKDVMKASVMLEHD-PQ 1030
Query 183 FAVVLGFDVKVD 194
+AV+L FDV+++
Sbjct 1031 YAVILAFDVRIE 1042
> YAL035w
Length=1002
Score = 112 bits (280), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 68/132 (51%), Positives = 92/132 (69%), Gaps = 1/132 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+R+K +Y H V+AA+GVKI A LE AV+G+ L V E+D EL +VM +++ +L
Sbjct 686 ELRLKSEYVHHKEVKAALGVKIAANDLEKAVSGSRLLVVGPEDDEDELMDDVMDDLTGLL 745
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
V+ T GV V ASTLG+LEALL +L+D KIPV ++ +G V K+D+ KAS M EK E
Sbjct 746 DSVDTTGKGVVVQASTLGSLEALLDFLKDMKIPVMSIGLGPVYKRDVMKASTMLEKA-PE 804
Query 183 FAVVLGFDVKVD 194
+AV+L FDVKVD
Sbjct 805 YAVMLCFDVKVD 816
> SPAC56F8.03
Length=1079
Score = 108 bits (270), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 66/132 (50%), Positives = 91/132 (68%), Gaps = 1/132 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
EMRVK Y H ++AAMGVKI A LE AVAG+ L V ++D +L E+M+++ +L
Sbjct 765 EMRVKSAYVHHKEIKAAMGVKICANDLEKAVAGSRLLVVGPDDDEEDLAEEIMEDLENLL 824
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
+++ + GV V ASTLG+LEALL +L+ KIPV +VNIG V KKD+ + + M EK E
Sbjct 825 GRIDTSGIGVSVQASTLGSLEALLEFLKQMKIPVASVNIGPVYKKDVMRCATMLEKA-KE 883
Query 183 FAVVLGFDVKVD 194
+A++L FDVKVD
Sbjct 884 YALMLCFDVKVD 895
> At2g27700
Length=479
Score = 108 bits (269), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 89/134 (66%), Gaps = 3/134 (2%)
Query 62 EEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLV 121
+E+ V G++ H ++AA + I+A LE + GT+L V ++DI ++ VM++V+ V
Sbjct 314 KELHVNGNHVHHEVIKAAECINIIAKDLEHVIVGTALHVVGPDDDIEAIKELVMEDVNSV 373
Query 122 LKKVEKTTNGVYVMASTLGALEALLAYLED--CKIPVFAVNIGTVQKKDIKKASVMREKG 179
L +++K+ GVY+ ASTLG+LEALL +L+ K+PV + IG VQKKD+ KA VM E+
Sbjct 374 LSRIDKSGEGVYIQASTLGSLEALLEFLKSPAVKLPVGGIGIGPVQKKDVMKAGVMLERK 433
Query 180 FAEFAVVLGFDVKV 193
EFA +L DV+V
Sbjct 434 -KEFATILALDVEV 446
> At1g76720
Length=1224
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 59/131 (45%), Positives = 82/131 (62%), Gaps = 8/131 (6%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVKG Y H ++AA G+KI A GLE A+AGTSL V ++DI ++ M+++ VL
Sbjct 940 ELRVKGTYLHHKEIKAAQGIKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVL 999
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFAE 182
+++K+ GVYV STLG+LEALL +L K P AVNI + + + EK E
Sbjct 1000 SRIDKSGEGVYVQTSTLGSLEALLEFL---KTP--AVNIPVSEGYNEGWSDAREEK---E 1051
Query 183 FAVVLGFDVKV 193
+A +L FDVKV
Sbjct 1052 YATILAFDVKV 1062
> ECU09g0070
Length=659
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 52/130 (40%), Positives = 85/130 (65%), Gaps = 3/130 (2%)
Query 62 EEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLV 121
+E+RVK Y V A++G+KI A GLE A+AG+ + + E E+ E++ + ++ V
Sbjct 348 KELRVKSQYIMVKEVRASLGIKIAAGGLEKAIAGSRVLIVEDNEE--EVKRALETDLESV 405
Query 122 LKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFA 181
+E + GV+V++STLG+LEAL+++L+ KIP+ +IGTV+KKD+ + M +K
Sbjct 406 FSSIELSQEGVHVVSSTLGSLEALISFLKTSKIPILGASIGTVKKKDMIIVASMAKK-HK 464
Query 182 EFAVVLGFDV 191
E+A +L FDV
Sbjct 465 EYAAILCFDV 474
> CE27011
Length=1173
Score = 76.3 bits (186), Expect = 4e-14, Method: Composition-based stats.
Identities = 46/133 (34%), Positives = 72/133 (54%), Gaps = 22/133 (16%)
Query 62 EEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLV 121
+E+RVK +Y + V+ A GVK++A LE +AG +++ ++E+++ L E ++++
Sbjct 885 KELRVKNEYIHYKEVKGARGVKVLAKNLEKVLAGLPIYITDREDEVDYLRHEADRQLANA 944
Query 122 LKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPVFAVNIGTVQKKDIKKASVMREKGFA 181
L + K G IP VNIG V KKD++KAS M+E A
Sbjct 945 LHAIRKKPEG---------------------NIPYSNVNIGPVHKKDVQKASAMKEHK-A 982
Query 182 EFAVVLGFDVKVD 194
E+A VL FDVKV+
Sbjct 983 EYACVLAFDVKVE 995
> 7292368
Length=932
Score = 75.5 bits (184), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 42/93 (45%), Positives = 58/93 (62%), Gaps = 0/93 (0%)
Query 63 EMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVL 122
E+RVK Y + V+AA GVKI A LE A+AG +L +A K +++ EV +E+ L
Sbjct 836 ELRVKNAYVEYKEVKAAQGVKIAAKDLEKAIAGINLLIAHKPDEVEICTEEVARELKSAL 895
Query 123 KKVEKTTNGVYVMASTLGALEALLAYLEDCKIP 155
++ GV+V ASTLG+LEALL +L KIP
Sbjct 896 SHIKLAQTGVHVQASTLGSLEALLEFLRTSKIP 928
> Hs22044172
Length=416
Score = 32.0 bits (71), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 43/84 (51%), Gaps = 13/84 (15%)
Query 13 KASLVIKRLAGSG-SGFAAAAAAATAAAAILLLLVVSVRGDSCSESYSYSEEMRVKGDYQ 71
+A++++K L G+G S + AA + +AAA+LL+ + VRG +C
Sbjct 324 QAAVLVKVLIGTGLSMWPAAGDQSLSAAAVLLMTAIIVRGTACPSKRQIPR--------- 374
Query 72 SHSFVEAAMGVKIVAPGLEDAVAG 95
SF+ A ++ ++ L D +AG
Sbjct 375 --SFINQANLIQRIS-SLGDGIAG 395
> CE12914
Length=329
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 54/122 (44%), Gaps = 7/122 (5%)
Query 73 HSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVSLVLKKVEKTTNGV 132
H +AA+ V A G +++ + E +AEL + Q+ + L +T N
Sbjct 74 HGNSDAALHVSSAATGWSNSIE----YFLEHGYTVAELYATSWQDTN-ALHAASRTHN-C 127
Query 133 YVMASTLGALEALLAYLEDCKIPVFAVNIG-TVQKKDIKKASVMREKGFAEFAVVLGFDV 191
+ LEA+LAY KI V ++G T+ +K IK SV G + + L V
Sbjct 128 KDLVRLRKFLEAVLAYTAAPKISVVTHSMGVTLARKIIKGGSVSASDGSCDLGLPLNKKV 187
Query 192 KV 193
+V
Sbjct 188 EV 189
> YPL110c_3
Length=401
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 43/87 (49%), Gaps = 1/87 (1%)
Query 103 KEEDIAELESEVMQEVSLVLKKVEKTTNGVYVMASTLGALEALLAYLEDCKIPV-FAVNI 161
+EE++ ++ E+ V VLK V NG ++ S+ ++ L+ IP+ F
Sbjct 248 EEEELGQIMMEMNHWVDTVLKVVFDNANGRDIIFSSFHPDICIMLSLKQPVIPILFLTEG 307
Query 162 GTVQKKDIKKASVMREKGFAEFAVVLG 188
G+ Q D++ +S+ FA+ +LG
Sbjct 308 GSEQMADLRASSLQNGIRFAKKWNLLG 334
> CE18030
Length=798
Score = 28.9 bits (63), Expect = 7.1, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 31/70 (44%), Gaps = 1/70 (1%)
Query 60 YSEEMRVKGDYQSHSFVEAAMGVKIVAPGLEDAVAGTSLFVAEKEEDIAELESEVMQEVS 119
Y+ K D + A+ KIV D+ S VA +E I E+E E ++
Sbjct 710 YARHQETKVDVKPKKAATPAVQKKIVPKVAADSSGDDSEEVASSDEGIQEVEEEQPKQKK 769
Query 120 LVLKKVEKTT 129
LV KKVE T
Sbjct 770 LV-KKVEPKT 778
Lambda K H
0.317 0.131 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264145066
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40