bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1826_orf1
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
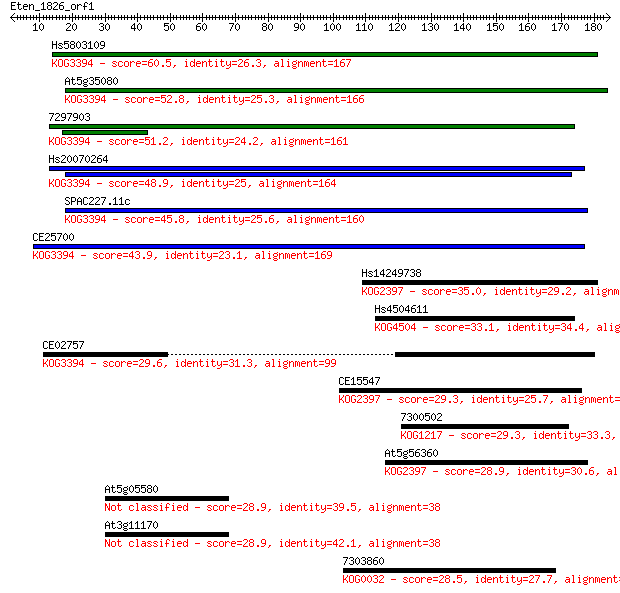
Hs5803109 60.5 2e-09
At5g35080 52.8 4e-07
7297903 51.2 1e-06
Hs20070264 48.9 6e-06
SPAC227.11c 45.8 5e-05
CE25700 43.9 2e-04
Hs14249738 35.0 0.094
Hs4504611 33.1 0.33
CE02757 29.6 3.9
CE15547 29.3 4.1
7300502 29.3 4.6
At5g56360 28.9 5.3
At5g05580 28.9 5.7
At3g11170 28.9 5.9
7303860 28.5 8.6
> Hs5803109
Length=667
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 44/167 (26%), Positives = 67/167 (40%), Gaps = 42/167 (25%)
Query 14 FTTDGWWSYEYCHPDSLVEYHKEPDGEVKGPLHLLGTLHASGDAAELIFKRPDPANPTLR 73
T WW+YE+C+ + +YH E D E+KG + LG ++ D
Sbjct 112 LKTKDWWTYEFCYGRHIQQYHME-DSEIKGEVLYLGYYQSAFD----------------- 153
Query 74 GTDTLPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCEGTDKQRSA 133
D +A+ R K + + NG+ C+ + R A
Sbjct 154 -WDDETAKASKQHRLK----------------------RYHSQTYGNGSKCDLNGRPREA 190
Query 134 RVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCPHPKLLPP 180
V F C G + + I +++E C+Y L I TP +CPHP L PP
Sbjct 191 EVRFLCDEGAGISGDY-IDRVDEPLSCSYVLTIRTPRLCPHPLLRPP 236
> At5g35080
Length=217
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 59/166 (35%), Gaps = 45/166 (27%)
Query 18 GWWSYEYCHPDSLVEYHKEPDGEVKGPLHLLGTLHASGDAAELIFKRPDPANPTLRGTDT 77
GWWSYE+CH + + H E + ++ LGT AA N T+
Sbjct 63 GWWSYEFCHQKYVRQLHVEDENKIVQEF-FLGTFDPEATAA---------FNQTV----- 107
Query 78 LPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCEGTDKQRSARVLF 137
S + T AS + + TNGT C+ T R V F
Sbjct 108 -------------------------SDASTDASQRYHSHVYTNGTTCDLTGSPREVEVRF 142
Query 138 ECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCPHPKLLPPRPL 183
C + + I E S C Y L + P +C HP +P+
Sbjct 143 VCAETRAM-----VTSITELSTCKYALTVQCPTLCKHPLFQLEKPV 183
> 7297903
Length=525
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/162 (24%), Positives = 69/162 (42%), Gaps = 31/162 (19%)
Query 13 TFTTDGWWSYEYCHPDSLVEYHKEPDGE-VKGPLHLLGTLHASGDAAELIFKRPDPANPT 71
T+ + +WSYE CH + +YH+E +G+ VK + LG K D
Sbjct 102 TYRIEAYWSYEICHGHHVRQYHEEREGKNVKFQEYYLG-------------KWTDEKMEI 148
Query 72 LRGTDTLPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCEGTDKQR 131
L +A P++K L ++ P ++ ++GT+C+ + R
Sbjct 149 LTKAWQADIKAGVKPKYKSLKIDNTRYP-------------YFEMEYSDGTMCDIINAPR 195
Query 132 SARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCP 173
+ V + C +P + I +ETS C Y+ +I + +CP
Sbjct 196 TTMVRYVC---YP-HGKNDIYSFKETSSCNYEAIILSSALCP 233
Score = 33.1 bits (74), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 17 DGWWSYEYCHPDSLVEYHKEPDGEVK 42
+GWW YE+C+ + ++HK+ EV+
Sbjct 361 NGWWKYEFCYGRHVRQFHKDKTSEVE 386
> Hs20070264
Length=483
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 41/167 (24%), Positives = 75/167 (44%), Gaps = 35/167 (20%)
Query 13 TFTTDGWWSYEYCHPDSLVEYHKEPDGEVKGPLH--LLGTLHASGDAAELIFKRPDPANP 70
++ + +W+YE CH + +YH+E + K +H LG + A L+F++ A
Sbjct 114 SYRIESYWTYEVCHGKHIRQYHEEKETGQKINIHEYYLGNMLAKN----LLFEKEREAEE 169
Query 71 TLRGTDTLPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCE-GTDK 129
+ ++ +P + + PV + NGT C ++
Sbjct 170 KEK-SNEIPTKNIEGQMTPYYPV-----------------------GMGNGTPCSLKQNR 205
Query 130 QRSARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCPHPK 176
RS+ V++ C P + H I+ + E + C Y+++I TPL+C HPK
Sbjct 206 PRSSTVMYIC---HPE-SKHEILSVAEVTTCEYEVVILTPLLCSHPK 248
Score = 35.8 bits (81), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 33/155 (21%), Positives = 57/155 (36%), Gaps = 37/155 (23%)
Query 18 GWWSYEYCHPDSLVEYHKEPDGEVKGPLHLLGTLHASGDAAELIFKRPDPANPTLRGTDT 77
GWW YE+C+ + +YH++ D SG + ++ GT
Sbjct 350 GWWKYEFCYGKHVHQYHEDKD---------------SGKTSVVV------------GTWN 382
Query 78 LPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCEGTDKQRSARVLF 137
A + + D Q + ++++ NG +C+ TDK R V
Sbjct 383 QEEHIEWAKKNTARAYHLQDDGTQ--------TVRMVSHFYGNGDICDITDKPRQVTVKL 434
Query 138 ECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVC 172
+C A V + E C Y L + +P++C
Sbjct 435 KCKESDSPHAV--TVYMLEPHSCQYILGVESPVIC 467
> SPAC227.11c
Length=322
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 41/164 (25%), Positives = 61/164 (37%), Gaps = 46/164 (28%)
Query 18 GWWSYEYCHPDSLVEYHKEP----DGEVKGPLHLLGTLHASGDAAELIFKRPDPANPTLR 73
G+W+Y+Y + + +YH EP D + P+++LGT P T +
Sbjct 132 GYWTYDYVYGQHVRQYHLEPQQGSDKVLANPMYILGTA---------------PNTQTKK 176
Query 74 GTDTLPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCEGTDKQRSA 133
+ N A F VE L NGT+C+ T + R
Sbjct 177 NLE-----ENWAIGF----VE---------------GKAYLQTTFRNGTMCDITKRPRHV 212
Query 134 RVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCPHPKL 177
+ +EC T I Q +E S C Y + IH P +C P
Sbjct 213 ILSYECSTNSD---TPEITQYQEVSSCAYSMTIHVPGLCSLPAF 253
> CE25700
Length=426
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/172 (22%), Positives = 70/172 (40%), Gaps = 44/172 (25%)
Query 8 EGWCSTFTTDGWWSYEYCHPDSLVEYHKEP--DGEVKGPLHLLGTLHASGDAAELIFKRP 65
+ CS + D +W+Y+ CH +++YH++ G+V LG ++ A+
Sbjct 106 DKMCS-YLIDVYWTYQVCHGRYVIQYHEDKMLTGQVSRTEFYLGNFDSALTAS-----TN 159
Query 66 DPANPTLRGTDTLPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCE 125
+ P R +E D P ++ +GT C+
Sbjct 160 EQVKPATR------------------RIENEDYP-------------YYSVSYNHGTSCD 188
Query 126 GTD-KQRSARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCPHPK 176
T K R+ V++ C H+I+ + E S C Y+++I T L+C HP+
Sbjct 189 VTGGKPRTTDVVYICVEK----VQHKILSVTEISSCHYEIVIMTDLLCKHPE 236
> Hs14249738
Length=305
Score = 35.0 bits (79), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 34/73 (46%), Gaps = 9/73 (12%)
Query 109 ASSKVLALQLTNGTVCEGTDKQRSARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHT 168
A++ + + +G C + R ++V C ++R+ + E S C Y L T
Sbjct 114 ANNTFTGMWMRDGDACR--SRSRQSKVELACG------KSNRLAHVSEPSTCVYALTFET 165
Query 169 PLVC-PHPKLLPP 180
PLVC PH L+ P
Sbjct 166 PLVCHPHALLVYP 178
> Hs4504611
Length=2491
Score = 33.1 bits (74), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 27/61 (44%), Gaps = 6/61 (9%)
Query 113 VLALQLTNGTVCEGTDKQRSARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVC 172
+L + T G C QRS + F C G T R V ++ETS C+Y T C
Sbjct 1308 LLKMNFTGGDTCHKV-YQRSTAIFFYCDRG-----TQRPVFLKETSDCSYLFEWRTQYAC 1361
Query 173 P 173
P
Sbjct 1362 P 1362
> CE02757
Length=821
Score = 29.6 bits (65), Expect = 3.9, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Query 11 CSTFTTDGWWSYEYCHPDSLVEYHKEP--DGEVKGPLHLL 48
C + WWSY C ++ + H EP +G VK L L
Sbjct 124 CVKLRGNHWWSYILCRGQTIEQVHGEPGQEGYVKNILGLF 163
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 28/63 (44%), Gaps = 3/63 (4%)
Query 119 TNGTVC--EGTDKQRSARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCPHPK 176
T+GT C E + R V +EC + I + E C Y +++ +C +P+
Sbjct 188 TSGTFCDLEEYREPRMTSVRYECDAQLSTNEVY-ISSVVEVKPCQYLMIVKVGTLCRYPE 246
Query 177 LLP 179
LP
Sbjct 247 FLP 249
> CE15547
Length=507
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 29/74 (39%), Gaps = 7/74 (9%)
Query 102 FSKSPTPASSKVLALQLTNGTVCEGTDKQRSARVLFECPVGFPVLATHRIVQIEETSFCT 161
F + P +K + +G C K RS + EC + +V++ E + C
Sbjct 432 FKEWSGPEGNKYSKMHFGDGQQCWNGPK-RSTDITIECG------EENELVEVTEPAKCE 484
Query 162 YDLLIHTPLVCPHP 175
Y TPL C P
Sbjct 485 YLFTFRTPLACADP 498
> 7300502
Length=833
Score = 29.3 bits (64), Expect = 4.6, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 23/51 (45%), Gaps = 1/51 (1%)
Query 121 GTVCEGTDKQRSARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLV 171
G C G + + L C F V H I+ TS CTY +I TP++
Sbjct 43 GRCCSGESDGATGKCLGSCKTRFRVCLKHYQATIDTTSQCTYGDVI-TPIL 92
> At5g56360
Length=647
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 29/63 (46%), Gaps = 9/63 (14%)
Query 116 LQLTNGTVC-EGTDKQRSARVLFECPVGFPVLATHRIVQIEETSFCTYDLLIHTPLVCPH 174
+ TNG C G D RS +V C + + ++ ++E S C Y ++ TP C
Sbjct 571 MSYTNGEKCWNGPD--RSLKVKLRCGL------KNELMDVDEPSRCEYAAILSTPARCLE 622
Query 175 PKL 177
KL
Sbjct 623 DKL 625
> At5g05580
Length=435
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 23/45 (51%), Gaps = 7/45 (15%)
Query 30 LVEYHKEPDGEVKGPLHLLGTL-------HASGDAAELIFKRPDP 67
L +Y++EP PLHLLG+L H D ++++ DP
Sbjct 384 LGKYYREPKNSGPLPLHLLGSLIKSMKQDHFVSDTGDVVYYEADP 428
> At3g11170
Length=446
Score = 28.9 bits (63), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 23/45 (51%), Gaps = 7/45 (15%)
Query 30 LVEYHKEPDGEVKGPLHLLGTL-------HASGDAAELIFKRPDP 67
L +Y++EPD PLHLL L H D E+++ + DP
Sbjct 391 LGKYYREPDKSGPLPLHLLEILAKSIKEDHYVSDEGEVVYYKADP 435
> 7303860
Length=762
Score = 28.5 bits (62), Expect = 8.6, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 30/65 (46%), Gaps = 2/65 (3%)
Query 103 SKSPTPASSKVLALQLTNGTVCEGTDKQRSARVLFECPVGFPVLATHRIVQIEETSFCTY 162
S + +S + +N + K R ++ + PVG P TH+ V +EE + T
Sbjct 632 SNPSSTGASNPITTSASNESASSRRAKFRINQMSRDVPVGLP--DTHQTVNLEEAANTTK 689
Query 163 DLLIH 167
D L+H
Sbjct 690 DCLLH 694
Lambda K H
0.319 0.138 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2950576972
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40