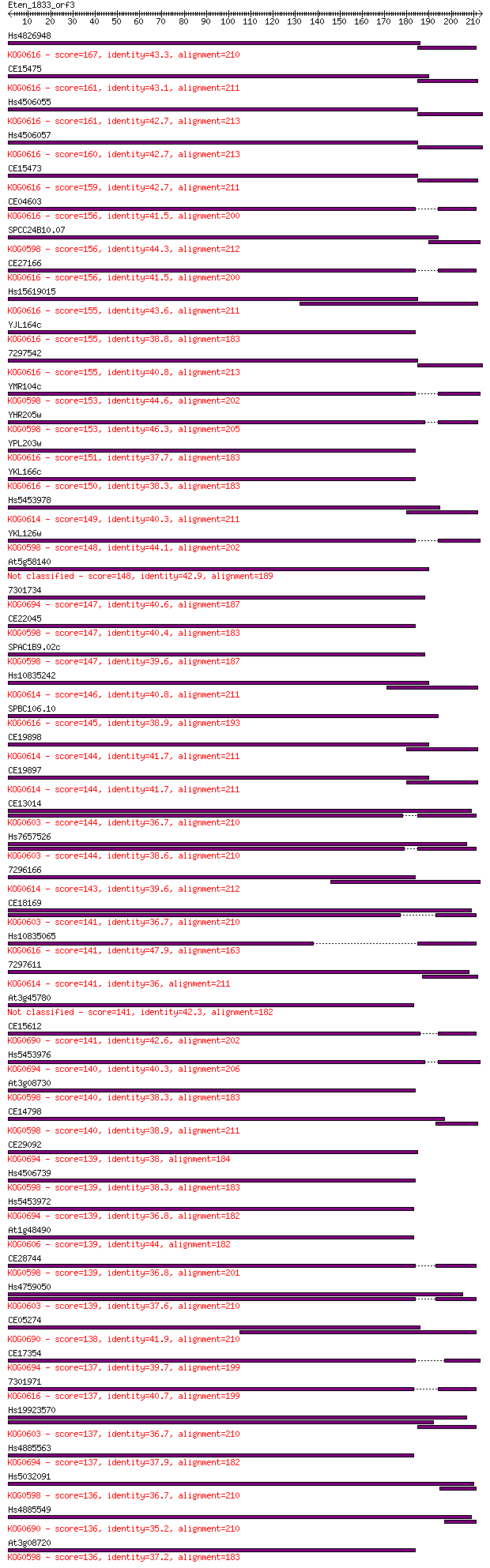
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1833_orf3
Length=213
Score E
Sequences producing significant alignments: (Bits) Value
Hs4826948 167 1e-41
CE15475 161 1e-39
Hs4506055 161 1e-39
Hs4506057 160 2e-39
CE15473 159 3e-39
CE04603 156 2e-38
SPCC24B10.07 156 3e-38
CE27166 156 3e-38
Hs15619015 155 5e-38
YJL164c 155 6e-38
7297542 155 8e-38
YMR104c 153 2e-37
YHR205w 153 2e-37
YPL203w 151 9e-37
YKL166c 150 2e-36
Hs5453978 149 3e-36
YKL126w 148 7e-36
At5g58140 148 9e-36
7301734 147 1e-35
CE22045 147 1e-35
SPAC1B9.02c 147 1e-35
Hs10835242 146 4e-35
SPBC106.10 145 6e-35
CE19898 144 1e-34
CE19897 144 1e-34
CE13014 144 2e-34
Hs7657526 144 2e-34
7296166 143 3e-34
CE18169 141 8e-34
Hs10835065 141 9e-34
7297611 141 1e-33
At3g45780 141 1e-33
CE15612 141 1e-33
Hs5453976 140 1e-33
At3g08730 140 2e-33
CE14798 140 2e-33
CE29092 139 3e-33
Hs4506739 139 3e-33
Hs5453972 139 4e-33
At1g48490 139 4e-33
CE28744 139 5e-33
Hs4759050 139 6e-33
CE05274 138 8e-33
CE17354 137 1e-32
7301971 137 1e-32
Hs19923570 137 2e-32
Hs4885563 137 2e-32
Hs5032091 136 3e-32
Hs4885549 136 3e-32
At3g08720 136 4e-32
> Hs4826948
Length=358
Score = 167 bits (424), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 78/186 (41%), Positives = 114/186 (61%), Gaps = 6/186 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L + LR R S F + E++ ++ L +++VYRDLK N+L+D DGH +L D
Sbjct 131 GGELFSYLRNRGRFSSTTGLFYSAEIICAIEYLHSKEIVYRDLKPENILLDRDGHIKLTD 190
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AKKL + RT++ CG+PEYL+PE++ GH +VD +ALG LI+E+L GFPPFF
Sbjct 191 FGFAKKL---VDRTWTLCGTPEYLAPEVIQSKGHGRAVDWWALGILIFEMLSGFPPFFDD 247
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
N +Y KI+ G++ FP + L+ +LL ++ R+G + G +V H WF
Sbjct 248 NPFGIYQKILAGKIDFPR--HLDFHVKDLIKKLLVVDRTRRLGNMKNGANDVKHHRWFRS 305
Query 180 VSWEAA 185
V WEA
Sbjct 306 VDWEAV 311
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 21/26 (80%), Gaps = 3/26 (11%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEML 210
AKKL + RT++ CG+PEYL+PE++
Sbjct 194 AKKL---VDRTWTLCGTPEYLAPEVI 216
> CE15475
Length=375
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 79/192 (41%), Positives = 119/192 (61%), Gaps = 8/192 (4%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + LR R SE +RF A +++L + L ++YRDLK N+LID G+ ++ D
Sbjct 134 GGEMFSHLRRIGRFSEPHSRFYAAQIVLAFEYLHSLDLIYRDLKPENLLIDSTGYLKITD 193
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ G RT++ CG+PEYL+PE++L G++ +VD +ALG LIYE+ G+PPFFA
Sbjct 194 FGFAKRVKG---RTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFAD 250
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+Y KI+ G++ FP+ S E + L+ LL ++ R G L GV ++ H WF
Sbjct 251 QPIQIYEKIVSGKVKFPS--HFSNELKDLLKNLLQVDLTKRYGNLKNGVADIKNHKWFGS 308
Query 180 VSWEA--AKKLT 189
W A KK+T
Sbjct 309 TDWIAIYQKKIT 320
Score = 35.0 bits (79), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 22/27 (81%), Gaps = 3/27 (11%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEMLL 211
AK++ G RT++ CG+PEYL+PE++L
Sbjct 197 AKRVKG---RTWTLCGTPEYLAPEIIL 220
> Hs4506055
Length=351
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 77/185 (41%), Positives = 117/185 (63%), Gaps = 6/185 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + LR R SE ARF A +++L + L ++YRDLK N+LID G+ ++ D
Sbjct 126 GGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTD 185
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ G RT++ CG+PEYL+PE++L G++ +VD +ALG LIYE+ G+PPFFA
Sbjct 186 FGFAKRVKG---RTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFAD 242
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+Y KI+ G++ FP+ S++ + L+ LL ++ R G L GV ++ H WFA
Sbjct 243 QPIQIYEKIVSGKVRFPSHF--SSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT 300
Query 180 VSWEA 184
W A
Sbjct 301 TDWIA 305
Score = 35.4 bits (80), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 22/29 (75%), Gaps = 3/29 (10%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEMLLGS 213
AK++ G RT++ CG+PEYL+PE++L
Sbjct 189 AKRVKG---RTWTLCGTPEYLAPEIILSK 214
> Hs4506057
Length=351
Score = 160 bits (405), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 77/185 (41%), Positives = 117/185 (63%), Gaps = 6/185 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + LR R SE ARF A +++L + L ++YRDLK N+LID G+ ++ D
Sbjct 126 GGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDHQGYIQVTD 185
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ G RT++ CG+PEYL+PE++L G++ +VD +ALG LIYE+ G+PPFFA
Sbjct 186 FGFAKRVKG---RTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFAD 242
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+Y KI+ G++ FP+ S++ + L+ LL ++ R G L GV ++ H WFA
Sbjct 243 QPIQIYEKIVSGKVRFPSHF--SSDLKDLLRNLLQVDLTKRFGNLKNGVSDIKTHKWFAT 300
Query 180 VSWEA 184
W A
Sbjct 301 TDWIA 305
Score = 35.4 bits (80), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 22/29 (75%), Gaps = 3/29 (10%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEMLLGS 213
AK++ G RT++ CG+PEYL+PE++L
Sbjct 189 AKRVKG---RTWTLCGTPEYLAPEIILSK 214
> CE15473
Length=359
Score = 159 bits (403), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 76/185 (41%), Positives = 115/185 (62%), Gaps = 6/185 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + LR R SE +RF A +++L + L ++YRDLK N+LID G+ ++ D
Sbjct 134 GGEMFSHLRRIGRFSEPHSRFYAAQIVLAFEYLHSLDLIYRDLKPENLLIDSTGYLKITD 193
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ G RT++ CG+PEYL+PE++L G++ +VD +ALG LIYE+ G+PPFFA
Sbjct 194 FGFAKRVKG---RTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFAD 250
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+Y KI+ G++ FP+ S E + L+ LL ++ R G L GV ++ H WF
Sbjct 251 QPIQIYEKIVSGKVKFPS--HFSNELKDLLKNLLQVDLTKRYGNLKNGVADIKNHKWFGS 308
Query 180 VSWEA 184
W A
Sbjct 309 TDWIA 313
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 22/27 (81%), Gaps = 3/27 (11%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEMLL 211
AK++ G RT++ CG+PEYL+PE++L
Sbjct 197 AKRVKG---RTWTLCGTPEYLAPEIIL 220
> CE04603
Length=371
Score = 156 bits (395), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 73/184 (39%), Positives = 116/184 (63%), Gaps = 6/184 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + LR + S + ARF A E++ L+ + +VYRDLK N+++ +GH ++AD
Sbjct 145 GGEMFSYLRASRSFSNSMARFYASEIVCALEYIHSLGIVYRDLKPENLMLSKEGHIKMAD 204
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK+L RT++ CG+P+YL+PE L +GH+ VD +ALG LIYE++VG PPF
Sbjct 205 FGFAKELRD---RTYTICGTPDYLAPESLARTGHNKGVDWWALGILIYEMMVGKPPFRGK 261
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+ +Y I++ +L FP S + A+ LV +LL ++ R+G + G ++V H WF
Sbjct 262 TTSEIYDAIIEHKLKFPRSFNLA--AKDLVKKLLEVDRTQRIGCMKNGTQDVKDHKWFEK 319
Query 180 VSWE 183
V+W+
Sbjct 320 VNWD 323
Score = 31.6 bits (70), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 10/17 (58%), Positives = 15/17 (88%), Gaps = 0/17 (0%)
Query 194 RTFSFCGSPEYLSPEML 210
RT++ CG+P+YL+PE L
Sbjct 214 RTYTICGTPDYLAPESL 230
> SPCC24B10.07
Length=569
Score = 156 bits (395), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 80/195 (41%), Positives = 116/195 (59%), Gaps = 5/195 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L+ A+F E+++ L+ L + V+YRDLK N+L+D GH L D
Sbjct 312 GGELFHHLQREGCFDTYRAKFYIAELLVALECLHEFNVIYRDLKPENILLDYTGHIALCD 371
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K + RT +FCG+PEYL+PE+LLG G+ VD + LG L+YE++ G PPF+
Sbjct 372 FGLCKLNMAKTDRTNTFCGTPEYLAPELLLGHGYTKVVDWWTLGVLLYEMITGLPPFYDE 431
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGV 180
N N MY KI+Q L FP + +A+ L+S LL+ P R+G+ GG +E+ H +F +
Sbjct 432 NINEMYRKILQDPLRFPDNID--EKAKDLLSGLLTRAPEKRLGS-GGAQEIKNHPFFDDI 488
Query 181 SWEA--AKKLTGQLK 193
W+ AKK+ K
Sbjct 489 DWKKLCAKKIQPPFK 503
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 190 GQLKRTFSFCGSPEYLSPEMLLG 212
+ RT +FCG+PEYL+PE+LLG
Sbjct 380 AKTDRTNTFCGTPEYLAPELLLG 402
> CE27166
Length=270
Score = 156 bits (394), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 73/184 (39%), Positives = 116/184 (63%), Gaps = 6/184 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + LR + S + ARF A E++ L+ + +VYRDLK N+++ +GH ++AD
Sbjct 44 GGEMFSYLRASRSFSNSMARFYASEIVCALEYIHSLGIVYRDLKPENLMLSKEGHIKMAD 103
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK+L RT++ CG+P+YL+PE L +GH+ VD +ALG LIYE++VG PPF
Sbjct 104 FGFAKELRD---RTYTICGTPDYLAPESLARTGHNKGVDWWALGILIYEMMVGKPPFRGK 160
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+ +Y I++ +L FP S + A+ LV +LL ++ R+G + G ++V H WF
Sbjct 161 TTSEIYDAIIEHKLKFPRSFNLA--AKDLVKKLLEVDRTQRIGCMKNGTQDVKDHKWFEK 218
Query 180 VSWE 183
V+W+
Sbjct 219 VNWD 222
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 10/17 (58%), Positives = 15/17 (88%), Gaps = 0/17 (0%)
Query 194 RTFSFCGSPEYLSPEML 210
RT++ CG+P+YL+PE L
Sbjct 113 RTYTICGTPDYLAPESL 129
> Hs15619015
Length=351
Score = 155 bits (393), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 78/185 (42%), Positives = 119/185 (64%), Gaps = 6/185 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + L+ R SE A F A +V+L +Q L +++RDLK N+LID G+ ++ D
Sbjct 126 GGEMFSRLQRVGRFSEPHACFYAAQVVLAVQYLHSLDLIHRDLKPENLLIDQQGYLQVTD 185
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ G RT++ CG+PEYL+PE++L G++ +VD +ALG LIYE+ VGFPPF+A
Sbjct 186 FGFAKRVKG---RTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAVGFPPFYAD 242
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+Y KI+ G++ FP+ S++ + L+ LL ++ R G L GV ++ H WFA
Sbjct 243 QPIQIYEKIVSGRVRFPSKL--SSDLKHLLRSLLQVDLTKRFGNLRNGVGDIKNHKWFAT 300
Query 180 VSWEA 184
SW A
Sbjct 301 TSWIA 305
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Query 132 GQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGVSWEAAKKLTGQ 191
G+ S P +C +A+ V L SL+ + R + + Q + + AK++ G
Sbjct 137 GRFSEPHACFYAAQVVLAVQYLHSLDLIHRDLKPENLL-IDQQGYLQVTDFGFAKRVKG- 194
Query 192 LKRTFSFCGSPEYLSPEMLL 211
RT++ CG+PEYL+PE++L
Sbjct 195 --RTWTLCGTPEYLAPEIIL 212
> YJL164c
Length=397
Score = 155 bits (392), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 71/184 (38%), Positives = 113/184 (61%), Gaps = 6/184 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L +LLR +R A+F A EV L L+ L + ++YRDLK N+L+D +GH ++ D
Sbjct 169 GGELFSLLRKSQRFPNPVAKFYAAEVCLALEYLHSKDIIYRDLKPENILLDKNGHIKITD 228
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK + T++ CG+P+Y++PE++ ++ S+D ++ G LIYE+L G+ PF+ +
Sbjct 229 FGFAKYVPD---VTYTLCGTPDYIAPEVVSTKPYNKSIDWWSFGILIYEMLAGYTPFYDS 285
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
N Y KI+ +L FP + + + L+SRL++ + R+G L G E+V H WF
Sbjct 286 NTMKTYEKILNAELRFPPFF--NEDVKDLLSRLITRDLSQRLGNLQNGTEDVKNHPWFKE 343
Query 180 VSWE 183
V WE
Sbjct 344 VVWE 347
> 7297542
Length=353
Score = 155 bits (391), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 73/185 (39%), Positives = 115/185 (62%), Gaps = 6/185 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ + LR R SE +RF A +++L + L ++YRDLK N+LID G+ ++ D
Sbjct 128 GGEMFSHLRKVGRFSEPHSRFYAAQIVLAFEYLHYLDLIYRDLKPENLLIDSQGYLKVTD 187
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ G RT++ CG+PEYL+PE++L G++ +VD +ALG L+YE+ G+PPFFA
Sbjct 188 FGFAKRVKG---RTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLVYEMAAGYPPFFAD 244
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
+Y KI+ G++ FP+ ++ + L+ LL ++ R G L GV ++ WFA
Sbjct 245 QPIQIYEKIVSGKVRFPS--HFGSDLKDLLRNLLQVDLTKRYGNLKAGVNDIKNQKWFAS 302
Query 180 VSWEA 184
W A
Sbjct 303 TDWIA 307
Score = 35.4 bits (80), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 22/29 (75%), Gaps = 3/29 (10%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEMLLGS 213
AK++ G RT++ CG+PEYL+PE++L
Sbjct 191 AKRVKG---RTWTLCGTPEYLAPEIILSK 216
> YMR104c
Length=677
Score = 153 bits (387), Expect = 2e-37, Method: Composition-based stats.
Identities = 77/183 (42%), Positives = 111/183 (60%), Gaps = 3/183 (1%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L+ R S A +RF E++ L L + V+YRDLK N+L+D GH L D
Sbjct 426 GGELFYHLQHEGRFSLARSRFYIAELLCALDSLHKLDVIYRDLKPENILLDYQGHIALCD 485
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K +T +FCG+PEYL+PE+LLG G+ +VD + LG L+YE++ G PP++
Sbjct 486 FGLCKLNMKDNDKTDTFCGTPEYLAPEILLGQGYTKTVDWWTLGILLYEMMTGLPPYYDE 545
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGV 180
N VMY KI+Q L FP + A+ L+ LLS +P R+G + G +E+ H +F +
Sbjct 546 NVPVMYKKILQQPLLFPDGFDPA--AKDLLIGLLSRDPSRRLG-VNGTDEIRNHPFFKDI 602
Query 181 SWE 183
SW+
Sbjct 603 SWK 605
Score = 37.0 bits (84), Expect = 0.033, Method: Composition-based stats.
Identities = 13/19 (68%), Positives = 18/19 (94%), Gaps = 0/19 (0%)
Query 194 RTFSFCGSPEYLSPEMLLG 212
+T +FCG+PEYL+PE+LLG
Sbjct 498 KTDTFCGTPEYLAPEILLG 516
> YHR205w
Length=824
Score = 153 bits (387), Expect = 2e-37, Method: Composition-based stats.
Identities = 83/189 (43%), Positives = 115/189 (60%), Gaps = 6/189 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L+ R SE A+F E++L L+ L +VYRDLK N+L+D +G+ L D
Sbjct 497 GGELFWHLQKEGRFSEDRAKFYIAELVLALEHLHDNDIVYRDLKPENILLDANGNIALCD 556
Query 61 FGLAKKLTGQLK-RTFSFCGSPEYLSPEMLLG-SGHDHSVDLYALGCLIYELLVGFPPFF 118
FGL+K LK RT +FCG+ EYL+PE+LL +G+ VD ++LG LI+E+ G+ PFF
Sbjct 557 FGLSK---ADLKDRTNTFCGTTEYLAPELLLDETGYTKMVDFWSLGVLIFEMCCGWSPFF 613
Query 119 AANRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFA 178
A N MY KI G++ FP S E RS V LL+ P R+GA+ E+ H +FA
Sbjct 614 AENNQKMYQKIAFGKVKFPRDV-LSQEGRSFVKGLLNRNPKHRLGAIDDGRELRAHPFFA 672
Query 179 GVSWEAAKK 187
+ WEA K+
Sbjct 673 DIDWEALKQ 681
Score = 32.7 bits (73), Expect = 0.50, Method: Composition-based stats.
Identities = 12/18 (66%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 194 RTFSFCGSPEYLSPEMLL 211
RT +FCG+ EYL+PE+LL
Sbjct 567 RTNTFCGTTEYLAPELLL 584
> YPL203w
Length=380
Score = 151 bits (382), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 69/184 (37%), Positives = 113/184 (61%), Gaps = 6/184 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L +LLR +R A+F A EV+L L+ L ++YRDLK N+L+D +GH ++ D
Sbjct 152 GGELFSLLRKSQRFPNPVAKFYAAEVILALEYLHAHNIIYRDLKPENILLDRNGHIKITD 211
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ T++ CG+P+Y++PE++ ++ SVD ++LG LIYE+L G+ PF+
Sbjct 212 FGFAKEVQ---TVTWTLCGTPDYIAPEVITTKPYNKSVDWWSLGVLIYEMLAGYTPFYDT 268
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
Y KI+QG++ +P + L+S+L++ + R+G L G ++ H WF+
Sbjct 269 TPMKTYEKILQGKVVYPPYFH--PDVVDLLSKLITADLTRRIGNLQSGSRDIKAHPWFSE 326
Query 180 VSWE 183
V WE
Sbjct 327 VVWE 330
> YKL166c
Length=398
Score = 150 bits (380), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 70/184 (38%), Positives = 111/184 (60%), Gaps = 6/184 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L +LLR +R A+F A EV L L+ L + ++YRDLK N+L+D +GH ++ D
Sbjct 170 GGELFSLLRKSQRFPNPVAKFYAAEVCLALEYLHSKDIIYRDLKPENILLDKNGHIKITD 229
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK + T++ CG+P+Y++PE++ ++ SVD ++ G LIYE+L G+ PF+ +
Sbjct 230 FGFAKYVPDV---TYTLCGTPDYIAPEVVSTKPYNKSVDWWSFGVLIYEMLAGYTPFYNS 286
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFAG 179
N Y I+ +L FP +A+ L+ +L++ + R+G L G E+V H WF
Sbjct 287 NTMKTYENILNAELKFPPFFH--PDAQDLLKKLITRDLSERLGNLQNGSEDVKNHPWFNE 344
Query 180 VSWE 183
V WE
Sbjct 345 VIWE 348
> Hs5453978
Length=762
Score = 149 bits (377), Expect = 3e-36, Method: Composition-based stats.
Identities = 75/199 (37%), Positives = 118/199 (59%), Gaps = 8/199 (4%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L ++LR E ++F V L + ++YRDLK N+++D +G+ +L D
Sbjct 535 GGELWSILRDRGSFDEPTSKFCVACVTEAFDYLHRLGIIYRDLKPENLILDAEGYLKLVD 594
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AKK+ G ++T++FCG+PEY++PE++L GHD SVD ++LG L+YELL G PPF
Sbjct 595 FGFAKKI-GSGQKTWTFCGTPEYVAPEVILNKGHDFSVDFWSLGILVYELLTGNPPFSGV 653
Query 121 NRNVMYMKIMQG--QLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWF 177
++ + Y I++G ++ FP E L+ RL P R+G L G+ ++ +H W
Sbjct 654 DQMMTYNLILKGIEKMDFPRKITRRPE--DLIRRLCRQNPTERLGNLKNGINDIKKHRWL 711
Query 178 AGVSWEA--AKKLTGQLKR 194
G +WE A+ L L+R
Sbjct 712 NGFNWEGLKARSLPSPLQR 730
Score = 36.6 bits (83), Expect = 0.036, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 26/32 (81%), Gaps = 1/32 (3%)
Query 180 VSWEAAKKLTGQLKRTFSFCGSPEYLSPEMLL 211
V + AKK+ G ++T++FCG+PEY++PE++L
Sbjct 593 VDFGFAKKI-GSGQKTWTFCGTPEYVAPEVIL 623
> YKL126w
Length=680
Score = 148 bits (374), Expect = 7e-36, Method: Composition-based stats.
Identities = 76/183 (41%), Positives = 109/183 (59%), Gaps = 3/183 (1%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L+ R + ARF E++ L L + VVYRDLK N+L+D GH L D
Sbjct 429 GGELFYHLQKEGRFDLSRARFYTAELLCALDNLHKLDVVYRDLKPENILLDYQGHIALCD 488
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K +T +FCG+PEYL+PE+LLG G+ +VD + LG L+YE+L G PP++
Sbjct 489 FGLCKLNMKDDDKTDTFCGTPEYLAPELLLGLGYTKAVDWWTLGVLLYEMLTGLPPYYDE 548
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGV 180
+ MY KI+Q L FP +A+ L+ LLS +P R+G G +E+ H +F+ +
Sbjct 549 DVPKMYKKILQEPLVFPDG--FDRDAKDLLIGLLSRDPTRRLG-YNGADEIRNHPFFSQL 605
Query 181 SWE 183
SW+
Sbjct 606 SWK 608
Score = 37.0 bits (84), Expect = 0.026, Method: Composition-based stats.
Identities = 13/19 (68%), Positives = 18/19 (94%), Gaps = 0/19 (0%)
Query 194 RTFSFCGSPEYLSPEMLLG 212
+T +FCG+PEYL+PE+LLG
Sbjct 501 KTDTFCGTPEYLAPELLLG 519
> At5g58140
Length=920
Score = 148 bits (373), Expect = 9e-36, Method: Composition-based stats.
Identities = 81/218 (37%), Positives = 123/218 (56%), Gaps = 29/218 (13%)
Query 1 GGDLAALL--RCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRL 58
GG+L ALL + K L+E +ARF A EV++GL+ L +VYRDLK N+L+ DGH L
Sbjct 664 GGELFALLDRQPMKILTEDSARFYAAEVVIGLEYLHCLGIVYRDLKPENILLKKDGHIVL 723
Query 59 ADFGLA------------------KKLTGQLKRTF---------SFCGSPEYLSPEMLLG 91
ADF L+ ++ Q TF SF G+ EY++PE++ G
Sbjct 724 ADFDLSFMTTCTPQLIIPAAPSKRRRSKSQPLPTFVAEPSTQSNSFVGTEEYIAPEIITG 783
Query 92 SGHDHSVDLYALGCLIYELLVGFPPFFAANRNVMYMKIMQGQLSFPASCQASAEARSLVS 151
+GH ++D +ALG L+YE+L G PF NR + I+ L+FP+S S R L++
Sbjct 784 AGHTSAIDWWALGILLYEMLYGRTPFRGKNRQKTFANILHKDLTFPSSIPVSLVGRQLIN 843
Query 152 RLLSLEPLMRVGALGGVEEVMQHSWFAGVSWEAAKKLT 189
LL+ +P R+G+ GG E+ QH++F G++W + ++
Sbjct 844 TLLNRDPSSRLGSKGGANEIKQHAFFRGINWPLIRGMS 881
> 7301734
Length=739
Score = 147 bits (372), Expect = 1e-35, Method: Composition-based stats.
Identities = 76/189 (40%), Positives = 114/189 (60%), Gaps = 6/189 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL ++ +R + A F A EV L LQ L V+YRDLK N+L+D +GHC+LAD
Sbjct 491 GGDLMFQIQKARRFEASRAAFYAAEVTLALQFLHTHGVIYRDLKLDNILLDQEGHCKLAD 550
Query 61 FGLAKK--LTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFF 118
FG+ K+ + G L T +FCG+P+Y++PE+L + SVD +ALG L+YE++ G PPF
Sbjct 551 FGMCKEGIMNGML--TTTFCGTPDYIAPEILKEQEYGASVDWWALGVLMYEMMAGQPPFE 608
Query 119 AANRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFA 178
A N + ++ IM + +P S EA S++ L+ P R+G G E+ +H +FA
Sbjct 609 ADNEDELFDSIMHDDVLYPV--WLSREAVSILKGFLTKNPEQRLGCTGDENEIRKHPFFA 666
Query 179 GVSWEAAKK 187
+ W+ +K
Sbjct 667 KLDWKELEK 675
> CE22045
Length=580
Score = 147 bits (372), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 74/184 (40%), Positives = 111/184 (60%), Gaps = 3/184 (1%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L E A+F E+++ L+ L QQ ++YRDLK N+L+D GH +L D
Sbjct 169 GGELFMHLEREGMFMENVAKFYLSEIVVSLEHLHQQGIIYRDLKPENILLDAYGHVKLTD 228
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K+ ++T +FCG+ EY++PE+L+ GH +VD ++LG L++++L G PPF A
Sbjct 229 FGLCKEEIEGDQKTHTFCGTIEYMAPEILMRCGHGKAVDWWSLGALMFDMLTGGPPFTAE 288
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWFAG 179
NR KI++G+L+ PA S EAR L+ +LL R+GA L EE+ H++F
Sbjct 289 NRRKTIDKILKGRLTLPAYL--SNEARDLIKKLLKRHVDTRLGAGLSDAEEIKSHAFFKT 346
Query 180 VSWE 183
W
Sbjct 347 TDWN 350
> SPAC1B9.02c
Length=696
Score = 147 bits (372), Expect = 1e-35, Method: Composition-based stats.
Identities = 74/188 (39%), Positives = 111/188 (59%), Gaps = 2/188 (1%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L+ R E A+F E++L L+ L + ++YRDLK N+L+D DGH L D
Sbjct 387 GGELFWHLQHEGRFPEQRAKFYIAELVLALEHLHKHDIIYRDLKPENILLDADGHIALCD 446
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLL-GSGHDHSVDLYALGCLIYELLVGFPPFFA 119
FGL+K T +FCG+ EYL+PE+LL G+ VD ++LG L++E+ G+ PF+A
Sbjct 447 FGLSKANLSANATTNTFCGTTEYLAPEVLLEDKGYTKQVDFWSLGVLVFEMCCGWSPFYA 506
Query 120 ANRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAG 179
+ MY I G++ FP S+E RS V LL+ P R+GA+ E+ +H +FA
Sbjct 507 PDVQQMYRNIAFGKVRFPKGV-LSSEGRSFVRGLLNRNPNHRLGAVADTTELKEHPFFAD 565
Query 180 VSWEAAKK 187
++W+ K
Sbjct 566 INWDLLSK 573
> Hs10835242
Length=686
Score = 146 bits (368), Expect = 4e-35, Method: Composition-based stats.
Identities = 74/192 (38%), Positives = 115/192 (59%), Gaps = 6/192 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L +LR ++ RF V+ L + ++YRDLK N+++D G+ +L D
Sbjct 458 GGELWTILRDRGSFEDSTTRFYTACVVEAFAYLHSKGIIYRDLKPENLILDHRGYAKLVD 517
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AKK+ G K+T++FCG+PEY++PE++L GHD S D ++LG L+YELL G PPF
Sbjct 518 FGFAKKI-GFGKKTWTFCGTPEYVAPEIILNKGHDISADYWSLGILMYELLTGSPPFSGP 576
Query 121 NRNVMYMKIMQG--QLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWF 177
+ Y I++G + FP + + A +L+ +L P R+G L GV+++ +H WF
Sbjct 577 DPMKTYNIILRGIDMIEFPK--KIAKNAANLIKKLCRDNPSERLGNLKNGVKDIQKHKWF 634
Query 178 AGVSWEAAKKLT 189
G +WE +K T
Sbjct 635 EGFNWEGLRKGT 646
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 31/42 (73%), Gaps = 2/42 (4%)
Query 171 VMQHSWFAG-VSWEAAKKLTGQLKRTFSFCGSPEYLSPEMLL 211
++ H +A V + AKK+ G K+T++FCG+PEY++PE++L
Sbjct 506 ILDHRGYAKLVDFGFAKKI-GFGKKTWTFCGTPEYVAPEIIL 546
> SPBC106.10
Length=512
Score = 145 bits (366), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 75/195 (38%), Positives = 122/195 (62%), Gaps = 9/195 (4%)
Query 1 GGDLAALLR-CHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLA 59
GG+L +LLR CH R E A+F A EV+L L L Q+VYRDLK N+L+D GH ++
Sbjct 283 GGELFSLLRKCH-RFPEKVAKFYAAEVILALDYLHHNQIVYRDLKPENLLLDRFGHLKIV 341
Query 60 DFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFA 119
DFG AK+++ + CG+P+YL+PE++ ++ + D ++LG LI+E+L G+PPF++
Sbjct 342 DFGFAKRVS--TSNCCTLCGTPDYLAPEIISLKPYNKAADWWSLGILIFEMLAGYPPFYS 399
Query 120 ANRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWFA 178
N +Y I++G++++P+ S + L+S LL + R G L G +++ H WF
Sbjct 400 ENPMKLYENILEGKVNYPSYF--SPASIDLLSHLLQRDITCRYGNLKDGSMDIIMHPWFR 457
Query 179 GVSWEAAKKLTGQLK 193
+SW+ K LT +++
Sbjct 458 DISWD--KILTRKIE 470
> CE19898
Length=737
Score = 144 bits (364), Expect = 1e-34, Method: Composition-based stats.
Identities = 77/192 (40%), Positives = 112/192 (58%), Gaps = 5/192 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L LR + ARF V+ GL+ L ++ +VYRDLK N L+ G+ +L D
Sbjct 509 GGELWTTLRDRGHFDDYTARFYVACVLEGLEYLHRKNIVYRDLKPENCLLANTGYLKLVD 568
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AKKL ++T++FCG+PEY+SPE++L GHD + D +ALG I EL++G PPF A+
Sbjct 569 FGFAKKLASG-RKTWTFCGTPEYVSPEIILNKGHDQAADYWALGIYICELMLGRPPFQAS 627
Query 121 NRNVMYMKIMQG--QLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWF 177
+ Y I++G L P + A +LV +L P R+G+ GGV ++ +H WF
Sbjct 628 DPMKTYTLILKGVDALEIPNR-RIGKTATALVKKLCRDNPGERLGSGSGGVNDIRKHRWF 686
Query 178 AGVSWEAAKKLT 189
G WE + T
Sbjct 687 MGFDWEGLRSRT 698
Score = 37.7 bits (86), Expect = 0.019, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 25/32 (78%), Gaps = 1/32 (3%)
Query 180 VSWEAAKKLTGQLKRTFSFCGSPEYLSPEMLL 211
V + AKKL ++T++FCG+PEY+SPE++L
Sbjct 567 VDFGFAKKLASG-RKTWTFCGTPEYVSPEIIL 597
> CE19897
Length=780
Score = 144 bits (364), Expect = 1e-34, Method: Composition-based stats.
Identities = 77/192 (40%), Positives = 112/192 (58%), Gaps = 5/192 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L LR + ARF V+ GL+ L ++ +VYRDLK N L+ G+ +L D
Sbjct 552 GGELWTTLRDRGHFDDYTARFYVACVLEGLEYLHRKNIVYRDLKPENCLLANTGYLKLVD 611
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AKKL ++T++FCG+PEY+SPE++L GHD + D +ALG I EL++G PPF A+
Sbjct 612 FGFAKKLASG-RKTWTFCGTPEYVSPEIILNKGHDQAADYWALGIYICELMLGRPPFQAS 670
Query 121 NRNVMYMKIMQG--QLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWF 177
+ Y I++G L P + A +LV +L P R+G+ GGV ++ +H WF
Sbjct 671 DPMKTYTLILKGVDALEIPNR-RIGKTATALVKKLCRDNPGERLGSGSGGVNDIRKHRWF 729
Query 178 AGVSWEAAKKLT 189
G WE + T
Sbjct 730 MGFDWEGLRSRT 741
Score = 37.7 bits (86), Expect = 0.019, Method: Composition-based stats.
Identities = 16/32 (50%), Positives = 25/32 (78%), Gaps = 1/32 (3%)
Query 180 VSWEAAKKLTGQLKRTFSFCGSPEYLSPEMLL 211
V + AKKL ++T++FCG+PEY+SPE++L
Sbjct 610 VDFGFAKKLASG-RKTWTFCGTPEYVSPEIIL 640
> CE13014
Length=740
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 77/211 (36%), Positives = 118/211 (55%), Gaps = 5/211 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL L +E +F E+ L L+ L +VYRDLK N+L+D DGH ++ D
Sbjct 131 GGDLFTRLSKEVMFTEDDVKFYLAELTLALEHLHSLGIVYRDLKPENILLDADGHIKVTD 190
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL+K+ K+T+SFCG+ EY++PE++ GH + D ++LG L++E+L G PF
Sbjct 191 FGLSKEAIDSEKKTYSFCGTVEYMAPEVINRRGHSMAADFWSLGVLMFEMLTGHLPFQGR 250
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWFAG 179
+RN +I++ +LS P + EA+SL+ L R+GA GVEE+ +H++FA
Sbjct 251 DRNDTMTQILKAKLSMPHFL--TQEAQSLLRALFKRNSQNRLGAGPDGVEEIKRHAFFAK 308
Query 180 VSWEA--AKKLTGQLKRTFSFCGSPEYLSPE 208
+ + K++ K S S Y PE
Sbjct 309 IDFVKLLNKEIDPPFKPALSTVDSTSYFDPE 339
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 60/187 (32%), Positives = 98/187 (52%), Gaps = 14/187 (7%)
Query 1 GGDLAALLRCHKRL-SEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDL-DGH--- 55
GG+L L K L SE + ++ +Q L QQV +RDL AAN+L L DG
Sbjct 473 GGELLDKLVNKKSLGSEKEVAAIMANLLNAVQYLHSQQVAHRDLTAANILFALKDGDPSS 532
Query 56 CRLADFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFP 115
R+ DFG AK+ + + C + ++++PE+L G+D S D+++LG L++ +L G
Sbjct 533 LRIVDFGFAKQSRAENGMLMTPCYTAQFVAPEVLRKQGYDRSCDVWSLGVLLHTMLTGCT 592
Query 116 PFFAANRNV---MYMKIMQGQLSF--PASCQASAEARSLVSRLLSLEPLMRVGALGGVEE 170
PF + + ++ G++S P S EA+ LV ++L ++P RV A ++
Sbjct 593 PFAMGPNDTPDQILQRVGDGKISMTHPVWDTISDEAKDLVRKMLDVDPNRRVTA----KQ 648
Query 171 VMQHSWF 177
+QH W
Sbjct 649 ALQHKWI 655
Score = 32.7 bits (73), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEML 210
+K+ K+T+SFCG+ EY++PE++
Sbjct 194 SKEAIDSEKKTYSFCGTVEYMAPEVI 219
> Hs7657526
Length=745
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 79/213 (37%), Positives = 117/213 (54%), Gaps = 10/213 (4%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGD+ L +E +F E+ L L L Q +VYRDLK N+L+D GH +L D
Sbjct 157 GGDVFTRLSKEVLFTEEDVKFYLAELALALDHLHQLGIVYRDLKPENILLDEIGHIKLTD 216
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL+K+ Q K+ +SFCG+ EY++PE++ GH S D ++ G L++E+L G PF
Sbjct 217 FGLSKESVDQEKKAYSFCGTVEYMAPEVVNRRGHSQSADWWSYGVLMFEMLTGTLPFQGK 276
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGV 180
+RN I++ +L P SAEA+SL+ L P R+G+ GVEE+ +H +FA +
Sbjct 277 DRNETMNMILKAKLGMPQFL--SAEAQSLLRMLFKRNPANRLGS-EGVEEIKRHLFFANI 333
Query 181 SWEAAKKLTGQL-------KRTFSFCGSPEYLS 206
W+ K Q K +FC PE+ +
Sbjct 334 DWDKLYKREVQPPFKPASGKPDDTFCFDPEFTA 366
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 56/188 (29%), Positives = 96/188 (51%), Gaps = 15/188 (7%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLD-----GH 55
GG+L + K SE A + + + L Q VV+RDLK +N+L +D
Sbjct 502 GGELLDRILKQKCFSEREASDILYVISKTVDYLHCQGVVHRDLKPSNILY-MDESASADS 560
Query 56 CRLADFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFP 115
R+ DFG AK+L G+ + C + +++PE+L+ G+D + D+++LG L Y +L G+
Sbjct 561 IRICDFGFAKQLRGENGLLLTPCYTANFVAPEVLMQQGYDAACDIWSLGVLFYTMLAGYT 620
Query 116 PFFAANRNV---MYMKIMQGQLSFPASC--QASAEARSLVSRLLSLEPLMRVGALGGVEE 170
PF + + ++I G+ S S A+ L+S +L ++P R A E+
Sbjct 621 PFANGPNDTPEEILLRIGNGKFSLSGGNWDNISDGAKDLLSHMLHMDPHQRYTA----EQ 676
Query 171 VMQHSWFA 178
+++HSW
Sbjct 677 ILKHSWIT 684
Score = 32.0 bits (71), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEML 210
+K+ Q K+ +SFCG+ EY++PE++
Sbjct 220 SKESVDQEKKAYSFCGTVEYMAPEVV 245
> 7296166
Length=768
Score = 143 bits (360), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 71/186 (38%), Positives = 114/186 (61%), Gaps = 6/186 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG++ +LR + AA+F+ V+ + L + ++YRDLK N+++D G+ ++ D
Sbjct 541 GGEIWTMLRDRGSFEDNAAQFIIGCVLQAFEYLHARGIIYRDLKPENLMLDERGYVKIVD 600
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK++ G +T++FCG+PEY++PE++L GHD +VD +ALG LI+ELL G PPF A
Sbjct 601 FGFAKQI-GTSSKTWTFCGTPEYVAPEIILNKGHDRAVDYWALGILIHELLNGTPPFSAP 659
Query 121 NRNVMYMKIMQG--QLSFPASCQASAEARSLVSRLLSLEPLMRVG-ALGGVEEVMQHSWF 177
+ Y I++G ++FP S A L+ RL P R+G GG++++ +H WF
Sbjct 660 DPMQTYNLILKGIDMIAFPK--HISRWAVQLIKRLCRDVPSERLGYQTGGIQDIKKHKWF 717
Query 178 AGVSWE 183
G W+
Sbjct 718 LGFDWD 723
Score = 37.0 bits (84), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 11/67 (16%)
Query 146 ARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGVSWEAAKKLTGQLKRTFSFCGSPEYL 205
AR ++ R L E LM + + + V + AK++ G +T++FCG+PEY+
Sbjct 575 ARGIIYRDLKPENLM----------LDERGYVKIVDFGFAKQI-GTSSKTWTFCGTPEYV 623
Query 206 SPEMLLG 212
+PE++L
Sbjct 624 APEIILN 630
> CE18169
Length=797
Score = 141 bits (356), Expect = 8e-34, Method: Composition-based stats.
Identities = 77/211 (36%), Positives = 118/211 (55%), Gaps = 5/211 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL L +E +F E+ L L+ L +VYRDLK N+L+D DGH ++ D
Sbjct 188 GGDLFTRLSKEVMFTEDDVKFYLAELTLALEHLHSLGIVYRDLKPENILLDADGHIKVTD 247
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL+K+ K+T+SFCG+ EY++PE++ GH + D ++LG L++E+L G PF
Sbjct 248 FGLSKEAIDSEKKTYSFCGTVEYMAPEVINRRGHSMAADFWSLGVLMFEMLTGHLPFQGR 307
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWFAG 179
+RN +I++ +LS P + EA+SL+ L R+GA GVEE+ +H++FA
Sbjct 308 DRNDTMTQILKAKLSMPHFL--TQEAQSLLRALFKRNSQNRLGAGPDGVEEIKRHAFFAK 365
Query 180 VSWEAA--KKLTGQLKRTFSFCGSPEYLSPE 208
+ + K++ K S S Y PE
Sbjct 366 IDFVKLLNKEIDPPFKPALSTVDSTSYFDPE 396
Score = 91.7 bits (226), Expect = 1e-18, Method: Composition-based stats.
Identities = 60/186 (32%), Positives = 98/186 (52%), Gaps = 14/186 (7%)
Query 1 GGDLAALLRCHKRL-SEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDL-DG---H 55
GG+L L K L SE + ++ +Q L QQV +RDL AAN+L L DG
Sbjct 530 GGELLDKLVNKKSLGSEKEVAAIMANLLNAVQYLHSQQVAHRDLTAANILFALKDGDPSS 589
Query 56 CRLADFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFP 115
R+ DFG AK+ + + C + ++++PE+L G+D S D+++LG L++ +L G
Sbjct 590 LRIVDFGFAKQSRAENGMLMTPCYTAQFVAPEVLRKQGYDRSCDVWSLGVLLHTMLTGCT 649
Query 116 PFFAANRNV---MYMKIMQGQLSF--PASCQASAEARSLVSRLLSLEPLMRVGALGGVEE 170
PF + + ++ G++S P S EA+ LV ++L ++P RV A ++
Sbjct 650 PFAMGPNDTPDQILQRVGDGKISMTHPVWDTISDEAKDLVRKMLDVDPNRRVTA----KQ 705
Query 171 VMQHSW 176
+QH W
Sbjct 706 ALQHKW 711
Score = 32.3 bits (72), Expect = 0.69, Method: Composition-based stats.
Identities = 10/18 (55%), Positives = 17/18 (94%), Gaps = 0/18 (0%)
Query 193 KRTFSFCGSPEYLSPEML 210
K+T+SFCG+ EY++PE++
Sbjct 259 KKTYSFCGTVEYMAPEVI 276
> Hs10835065
Length=277
Score = 141 bits (356), Expect = 9e-34, Method: Compositional matrix adjust.
Identities = 64/137 (46%), Positives = 91/137 (66%), Gaps = 3/137 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L + LR S F + E++ ++ L +++VYRDLK N+L+D DGH +L D
Sbjct 131 GGELFSYLRNRGHFSSTTGLFYSAEIICAIEYLHSKEIVYRDLKPENILLDRDGHIKLTD 190
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AKKL + RT++ CG+PEYL+PE++ GH +VD +ALG LI+E+L GFPPFF
Sbjct 191 FGFAKKL---VDRTWTLCGTPEYLAPEVIQSKGHGRAVDWWALGILIFEMLSGFPPFFDD 247
Query 121 NRNVMYMKIMQGQLSFP 137
N +Y KI+ G+L FP
Sbjct 248 NPFGIYQKILAGKLYFP 264
Score = 33.5 bits (75), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 21/26 (80%), Gaps = 3/26 (11%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEML 210
AKKL + RT++ CG+PEYL+PE++
Sbjct 194 AKKL---VDRTWTLCGTPEYLAPEVI 216
> 7297611
Length=1003
Score = 141 bits (355), Expect = 1e-33, Method: Composition-based stats.
Identities = 74/210 (35%), Positives = 114/210 (54%), Gaps = 4/210 (1%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGD+ ++ + E A+F+A V+ L +YRDLK N+++ DG+C+L D
Sbjct 775 GGDVWTVMSKRQYFDEKTAKFIAGCVVEAFDYLHSHHFIYRDLKPENLMLGTDGYCKLVD 834
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AK Q ++T +F G+PEY++PE++L GHD +VD +ALG L+YELLVG PF
Sbjct 835 FGFAK-FVRQNEKTNTFAGTPEYVAPEIILDRGHDRAVDYWALGILVYELLVGKTPFRGV 893
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVG-ALGGVEEVMQHSWFAG 179
N+ +Y +I+ G + A+ LV L P R+G G+ ++ +HSWF
Sbjct 894 NQIKIYQQILSGIDVIHMPSRIPKSAQHLVRHLCKQLPAERLGYQRKGIADIKRHSWFES 953
Query 180 VSWE--AAKKLTGQLKRTFSFCGSPEYLSP 207
+ W+ K+L +KR +Y P
Sbjct 954 LDWQRLKLKQLPSPIKRPLKSWTDLQYFGP 983
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 187 KLTGQLKRTFSFCGSPEYLSPEMLL 211
K Q ++T +F G+PEY++PE++L
Sbjct 839 KFVRQNEKTNTFAGTPEYVAPEIIL 863
> At3g45780
Length=996
Score = 141 bits (355), Expect = 1e-33, Method: Composition-based stats.
Identities = 77/213 (36%), Positives = 114/213 (53%), Gaps = 31/213 (14%)
Query 1 GGDLAALLRCHKR--LSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRL 58
GG+L LL R L E A RF A +V++ L+ L Q ++YRDLK NVLI +G L
Sbjct 745 GGELFMLLDRQPRKVLKEDAVRFYAAQVVVALEYLHCQGIIYRDLKPENVLIQGNGDISL 804
Query 59 ADFGLAKKLTGQ-----------------------------LKRTFSFCGSPEYLSPEML 89
+DF L+ + + ++ + SF G+ EY++PE++
Sbjct 805 SDFDLSCLTSCKPQLLIPSIDEKKKKKQQKSQQTPIFMAEPMRASNSFVGTEEYIAPEII 864
Query 90 LGSGHDHSVDLYALGCLIYELLVGFPPFFAANRNVMYMKIMQGQLSFPASCQASAEARSL 149
G+GH +VD +ALG L+YE+L G+ PF R + ++Q L FPAS AS + + L
Sbjct 865 SGAGHTSAVDWWALGILMYEMLYGYTPFRGKTRQKTFTNVLQKDLKFPASIPASLQVKQL 924
Query 150 VSRLLSLEPLMRVGALGGVEEVMQHSWFAGVSW 182
+ RLL +P R+G G EV QHS+F G++W
Sbjct 925 IFRLLQRDPKKRLGCFEGANEVKQHSFFKGINW 957
> CE15612
Length=541
Score = 141 bits (355), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 75/188 (39%), Positives = 114/188 (60%), Gaps = 7/188 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L +R SE ARF E++L L L + +VYRD+K N+L+D DGH ++AD
Sbjct 275 GGELFTHVRKCGTFSEPRARFYGAEIVLALGYLHRCDIVYRDMKLENLLLDKDGHIKIAD 334
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K+ +T +FCG+PEYL+PE+L + VD + +G ++YE++ G PF++
Sbjct 335 FGLCKEEISFGDKTSTFCGTPEYLAPEVLDDHDYGRCVDWWGVGVVMYEMMCGRLPFYSK 394
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVE---EVMQHSWF 177
+ N ++ IM G L FP+ + S EAR+L++ LL +P R+G GG E E+ + +F
Sbjct 395 DHNKLFELIMAGDLRFPS--KLSQEARTLLTGLLVKDPTQRLG--GGPEDALEICRADFF 450
Query 178 AGVSWEAA 185
V WEA
Sbjct 451 RTVDWEAT 458
Score = 32.0 bits (71), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 11/17 (64%), Positives = 16/17 (94%), Gaps = 0/17 (0%)
Query 194 RTFSFCGSPEYLSPEML 210
+T +FCG+PEYL+PE+L
Sbjct 347 KTSTFCGTPEYLAPEVL 363
> Hs5453976
Length=706
Score = 140 bits (354), Expect = 1e-33, Method: Composition-based stats.
Identities = 72/188 (38%), Positives = 114/188 (60%), Gaps = 7/188 (3%)
Query 1 GGDLAALLR-CHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLA 59
GGDL ++ CHK + A F A E++LGLQ L + +VYRDLK N+L+D DGH ++A
Sbjct 463 GGDLMYHIQSCHK-FDLSRATFYAAEIILGLQFLHSKGIVYRDLKLDNILLDKDGHIKIA 521
Query 60 DFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFA 119
DFG+ K+ +T +FCG+P+Y++PE+LLG ++HSVD ++ G L+YE+L+G PF
Sbjct 522 DFGMCKENMLGDAKTNTFCGTPDYIAPEILLGQKYNHSVDWWSFGVLLYEMLIGQSPFHG 581
Query 120 ANRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAG 179
+ ++ I +P + EA+ L+ +L EP R+G G ++ QH F
Sbjct 582 QDEEELFHSIRMDNPFYPRWLE--KEAKDLLVKLFVREPEKRLGVRG---DIRQHPLFRE 636
Query 180 VSWEAAKK 187
++WE ++
Sbjct 637 INWEELER 644
Score = 34.7 bits (78), Expect = 0.15, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 18/19 (94%), Gaps = 0/19 (0%)
Query 194 RTFSFCGSPEYLSPEMLLG 212
+T +FCG+P+Y++PE+LLG
Sbjct 535 KTNTFCGTPDYIAPEILLG 553
> At3g08730
Length=465
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 70/184 (38%), Positives = 109/184 (59%), Gaps = 5/184 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG L L E AR E++ + L ++ +++RDLK N+L+D DGH L D
Sbjct 216 GGHLFFQLYHQGLFREDLARVYTAEIVSAVSHLHEKGIMHRDLKPENILMDTDGHVMLTD 275
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGLAK+ + R+ S CG+ EY++PE++ G GHD + D +++G L+YE+L G PPF +
Sbjct 276 FGLAKEFE-ENTRSNSMCGTTEYMAPEIVRGKGHDKAADWWSVGILLYEMLTGKPPFLGS 334
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWFAG 179
+ KI++ ++ P S EA +++ LL EP R+G+ L G EE+ QH WF G
Sbjct 335 KGKIQ-QKIVKDKIKLPQFL--SNEAHAILKGLLQKEPERRLGSGLSGAEEIKQHKWFKG 391
Query 180 VSWE 183
++W+
Sbjct 392 INWK 395
> CE14798
Length=422
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 72/196 (36%), Positives = 112/196 (57%), Gaps = 4/196 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L + L+ K SE+ +RF A E+ L L ++ ++YRDLK N+L+D G+ L D
Sbjct 177 GGELFSHLQREKHFSESRSRFYAAEIACALGYLHEKNIIYRDLKPENLLLDDKGYLVLTD 236
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K+ K T +FCG+PEYL+PE++L +D +VD + LG ++YE++ G PPF++
Sbjct 237 FGLCKEDMQGSKTTSTFCGTPEYLAPEIILKKPYDKTVDWWCLGSVLYEMIFGLPPFYSK 296
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGV 180
+ N MY KI+ L + S L++ LL + R+G ++ H +F V
Sbjct 297 DHNEMYDKIINQPLRLKHNI--SVPCSELITGLLQKDRSKRLGHRNDFRDIRDHPFFLPV 354
Query 181 SWEAAKKLTGQLKRTF 196
W+ K L +LK F
Sbjct 355 DWD--KLLNRELKAPF 368
Score = 33.9 bits (76), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 17/19 (89%), Gaps = 0/19 (0%)
Query 193 KRTFSFCGSPEYLSPEMLL 211
K T +FCG+PEYL+PE++L
Sbjct 248 KTTSTFCGTPEYLAPEIIL 266
> CE29092
Length=707
Score = 139 bits (351), Expect = 3e-33, Method: Composition-based stats.
Identities = 70/187 (37%), Positives = 112/187 (59%), Gaps = 5/187 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL ++ ++ E+ ARF A EV LQ L + V+YRDLK N+L+D +GHCRLAD
Sbjct 461 GGDLMFQIQRARKFDESRARFYAAEVTCALQFLHRNDVIYRDLKLDNILLDAEGHCRLAD 520
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG+ K+ + T +FCG+P+Y++PE+L + SVD +ALG L+YE++ G PPF A
Sbjct 521 FGMCKEGINKDNLTSTFCGTPDYIAPEILQEMEYGVSVDWWALGVLMYEMMAGQPPFEAD 580
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL---GGVEEVMQHSWF 177
N + ++ I+ + +P S EA +++ ++ R+G + GG + + H +F
Sbjct 581 NEDDLFEAILNDDVLYPV--WLSKEAVNILKAFMTKNAGKRLGCVVSQGGEDAIRAHPFF 638
Query 178 AGVSWEA 184
+ W+A
Sbjct 639 REIDWDA 645
> Hs4506739
Length=495
Score = 139 bits (351), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 70/184 (38%), Positives = 107/184 (58%), Gaps = 3/184 (1%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L E A F E+ L L L Q ++YRDLK N+++ GH +L D
Sbjct 166 GGELFTHLEREGIFLEDTACFYLAEITLALGHLHSQGIIYRDLKPENIMLSSQGHIKLTD 225
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K+ + T +FCG+ EY++PE+L+ SGH+ +VD ++LG L+Y++L G PPF A
Sbjct 226 FGLCKESIHEGAVTHTFCGTIEYMAPEILVRSGHNRAVDWWSLGALMYDMLTGSPPFTAE 285
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRV-GALGGVEEVMQHSWFAG 179
NR KI++G+L+ P + +AR LV + L P R+ G G +V +H +F
Sbjct 286 NRKKTMDKIIRGKLALPPYL--TPDARDLVKKFLKRNPSQRIGGGPGDAADVQRHPFFRH 343
Query 180 VSWE 183
++W+
Sbjct 344 MNWD 347
> Hs5453972
Length=682
Score = 139 bits (350), Expect = 4e-33, Method: Composition-based stats.
Identities = 67/184 (36%), Positives = 111/184 (60%), Gaps = 4/184 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL ++ +R EA ARF A E++ L L + ++YRDLK NVL+D +GHC+LAD
Sbjct 437 GGDLMFHIQKSRRFDEARARFYAAEIISALMFLHDKGIIYRDLKLDNVLLDHEGHCKLAD 496
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG+ K+ T +FCG+P+Y++PE+L + +VD +A+G L+YE+L G PF A
Sbjct 497 FGMCKEGICNGVTTATFCGTPDYIAPEILQEMLYGPAVDWWAMGVLLYEMLCGHAPFEAE 556
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL--GGVEEVMQHSWFA 178
N + ++ I+ ++ +P +A ++ ++ P MR+G+L GG +++H +F
Sbjct 557 NEDDLFEAILNDEVVYPTWLH--EDATGILKSFMTKNPTMRLGSLTQGGEHAILRHPFFK 614
Query 179 GVSW 182
+ W
Sbjct 615 EIDW 618
> At1g48490
Length=1244
Score = 139 bits (350), Expect = 4e-33, Method: Composition-based stats.
Identities = 80/208 (38%), Positives = 115/208 (55%), Gaps = 27/208 (12%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGD ++LR L EA AR EV+L L+ L + VV+RDLK N+LI DGH +L D
Sbjct 910 GGDFYSMLRKIGCLDEANARVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHVKLTD 969
Query 61 FGLAK--------KLTGQLKRTFSF-----------------CGSPEYLSPEMLLGSGHD 95
FGL+K L+G + S G+P+YL+PE+LLG+GH
Sbjct 970 FGLSKVGLINNTDDLSGPVSSATSLLVEEKPKLPTLDHKRSAVGTPDYLAPEILLGTGHG 1029
Query 96 HSVDLYALGCLIYELLVGFPPFFAANRNVMYMKIMQGQLSFPASCQ-ASAEARSLVSRLL 154
+ D +++G ++YE LVG PPF A + ++ I+ + +P + S EAR L+ RLL
Sbjct 1030 ATADWWSVGIILYEFLVGIPPFNADHPQQIFDNILNRNIQWPPVPEDMSHEARDLIDRLL 1089
Query 155 SLEPLMRVGALGGVEEVMQHSWFAGVSW 182
+ +P R+GA G EV QHS+F + W
Sbjct 1090 TEDPHQRLGARGAA-EVKQHSFFKDIDW 1116
> CE28744
Length=879
Score = 139 bits (349), Expect = 5e-33, Method: Composition-based stats.
Identities = 65/183 (35%), Positives = 108/183 (59%), Gaps = 6/183 (3%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L LL E ARF E++L ++ L +VYRDLK NV+++L GH L D
Sbjct 109 GGELYTLLERKSDWPEEYARFYLSEIILAIEHLHGHDIVYRDLKPDNVMLNLAGHVVLTD 168
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K + ++T +FCG+ EY++PEM+ GHDH+VD++ALG L+Y++ +G PPF
Sbjct 169 FGLCKYKLKKGEKTLTFCGTHEYMAPEMIRKVGHDHAVDVWALGILMYDMFMGGPPFTGE 228
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGV 180
I++G++ P + SA + L+ R++ +P +R+ + ++ +H +F +
Sbjct 229 TPADKDKSILKGKVRLPP--KLSASGKDLIKRIIKRDPTLRI----TIPDIKEHEFFEEI 282
Query 181 SWE 183
W+
Sbjct 283 DWD 285
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 9/18 (50%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 193 KRTFSFCGSPEYLSPEML 210
++T +FCG+ EY++PEM+
Sbjct 180 EKTLTFCGTHEYMAPEMI 197
> Hs4759050
Length=740
Score = 139 bits (349), Expect = 6e-33, Method: Composition-based stats.
Identities = 77/214 (35%), Positives = 115/214 (53%), Gaps = 14/214 (6%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL L +E +F E+ L L L ++YRDLK N+L+D +GH +L D
Sbjct 152 GGDLFTRLSKEVMFTEEDVKFYLAELALALDHLHSLGIIYRDLKPENILLDEEGHIKLTD 211
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL+K+ K+ +SFCG+ EY++PE++ GH S D ++ G L++E+L G PF
Sbjct 212 FGLSKESIDHEKKAYSFCGTVEYMAPEVVNRRGHTQSADWWSFGVLMFEMLTGTLPFQGK 271
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWFAG 179
+R I++ +L P S EA+SL+ L P R+GA GVEE+ +HS+F+
Sbjct 272 DRKETMTMILKAKLGMPQFL--SPEAQSLLRMLFKRNPANRLGAGPDGVEEIKRHSFFST 329
Query 180 VSWEAA---------KKLTGQLKRTFSFCGSPEY 204
+ W K TG+ + TF F PE+
Sbjct 330 IDWNKLYRREIHPPFKPATGRPEDTFYF--DPEF 361
Score = 88.6 bits (218), Expect = 9e-18, Method: Composition-based stats.
Identities = 59/192 (30%), Positives = 99/192 (51%), Gaps = 15/192 (7%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVL-IDLDGH---C 56
GG+L + K SE A V + ++ L Q VV+RDLK +N+L +D G+
Sbjct 498 GGELLDKILRQKFFSEREASAVLFTITKTVEYLHAQGVVHRDLKPSNILYVDESGNPESI 557
Query 57 RLADFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPP 116
R+ DFG AK+L + + C + +++PE+L G+D + D+++LG L+Y +L G+ P
Sbjct 558 RICDFGFAKQLRAENGLLMTPCYTANFVAPEVLKRQGYDAACDIWSLGVLLYTMLTGYTP 617
Query 117 FFAANRNV---MYMKIMQGQLSFPASC--QASAEARSLVSRLLSLEPLMRVGALGGVEEV 171
F + + +I G+ S S A+ LVS++L ++P R+ A V
Sbjct 618 FANGPDDTPEEILARIGSGKFSLSGGYWNSVSDTAKDLVSKMLHVDPHQRLTA----ALV 673
Query 172 MQHSWFAGVSWE 183
++H W V W+
Sbjct 674 LRHPWI--VHWD 683
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 9/18 (50%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 193 KRTFSFCGSPEYLSPEML 210
K+ +SFCG+ EY++PE++
Sbjct 223 KKAYSFCGTVEYMAPEVV 240
> CE05274
Length=546
Score = 138 bits (348), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 77/193 (39%), Positives = 114/193 (59%), Gaps = 12/193 (6%)
Query 1 GGDLAALLRCHKRL-----SEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGH 55
GGDL L ++ SE ARF E++L L L +VYRDLK N+L+D DGH
Sbjct 275 GGDLYYHLNREVQMNKEGFSEPRARFYGSEIVLALGYLHANSIVYRDLKLENLLLDKDGH 334
Query 56 CRLADFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFP 115
++ADFGL K+ +T +FCG+PEYL+PE+L + VD + +G ++YE++ G
Sbjct 335 IKIADFGLCKEEISFGDKTSTFCGTPEYLAPEVLDDHDYGRCVDWWGVGVVMYEMMCGRL 394
Query 116 PFFAANRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVE---EVM 172
PF++ + N ++ IM G L FP+ + S EAR+L++ LL +P R+G GG E E+
Sbjct 395 PFYSKDHNKLFELIMAGDLRFPS--KLSQEARTLLTGLLVKDPTQRLG--GGPEDALEIC 450
Query 173 QHSWFAGVSWEAA 185
+ +F V WEA
Sbjct 451 RADFFRTVDWEAT 463
Score = 32.3 bits (72), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 48/119 (40%), Gaps = 30/119 (25%)
Query 105 CLIYELLVGFPPFFAANRNVMYMK--IMQGQLSFPASCQASA----EARSLVSRLLSLEP 158
C + E +G ++ NR V K + + F S A A S+V R L LE
Sbjct 267 CFVMEFAIGGDLYYHLNREVQMNKEGFSEPRARFYGSEIVLALGYLHANSIVYRDLKLEN 326
Query 159 LM-------RVGALGGVEEVMQHSWFAGVSWEAAKKLTGQLKRTFSFCGSPEYLSPEML 210
L+ ++ G +E + +T +FCG+PEYL+PE+L
Sbjct 327 LLLDKDGHIKIADFGLCKEEISFG-----------------DKTSTFCGTPEYLAPEVL 368
> CE17354
Length=567
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 70/185 (37%), Positives = 109/185 (58%), Gaps = 4/185 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL ++ K+ EA RF A E+++ LQ L ++YRDLK NVL+D DGH +LAD
Sbjct 321 GGDLMHHIQQIKKFDEARTRFYACEIVVALQFLHTNNIIYRDLKLDNVLLDCDGHIKLAD 380
Query 61 FGLAK-KLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFA 119
FG+AK ++ + +FCG+P+Y+SPE++ G ++ +VD ++ G L+YE+LVG PF
Sbjct 381 FGMAKTEMNRENGMASTFCGTPDYISPEIIKGQLYNEAVDFWSFGVLMYEMLVGQSPFHG 440
Query 120 ANRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVE-EVMQHSWFA 178
+ ++ I+ + FP + S EA +S L P R+G + + QH +F
Sbjct 441 EGEDELFDSILNERPYFPKT--ISKEAAKCLSALFDRNPNTRLGMPECPDGPIRQHCFFR 498
Query 179 GVSWE 183
GV W+
Sbjct 499 GVDWK 503
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 9/16 (56%), Positives = 15/16 (93%), Gaps = 0/16 (0%)
Query 197 SFCGSPEYLSPEMLLG 212
+FCG+P+Y+SPE++ G
Sbjct 397 TFCGTPDYISPEIIKG 412
> 7301971
Length=356
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 71/185 (38%), Positives = 104/185 (56%), Gaps = 8/185 (4%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L R ++ +E ARF A +V L L+ L ++YRDLK N+++D +G+ ++ D
Sbjct 129 GGELFTYHRKVRKFTEKQARFYAAQVFLALEYLHHCSLLYRDLKPENIMMDKNGYLKVTD 188
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG AKK+ RT + CG+PEYL PE++ + SVD +A G L++E + G PF A
Sbjct 189 FGFAKKVET---RTMTLCGTPEYLPPEIIQSKPYGTSVDWWAFGVLVFEFVAGHSPFSAH 245
Query 121 NRNV--MYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGAL-GGVEEVMQHSWF 177
NR+V MY KI + P+ S R LV LL ++ R G L G ++ +H WF
Sbjct 246 NRDVMSMYNKICEADYKMPSYF--SGALRHLVDHLLQVDLSKRFGNLINGNRDIKEHEWF 303
Query 178 AGVSW 182
V W
Sbjct 304 KDVEW 308
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 10/17 (58%), Positives = 14/17 (82%), Gaps = 0/17 (0%)
Query 194 RTFSFCGSPEYLSPEML 210
RT + CG+PEYL PE++
Sbjct 198 RTMTLCGTPEYLPPEII 214
> Hs19923570
Length=733
Score = 137 bits (345), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 75/216 (34%), Positives = 114/216 (52%), Gaps = 14/216 (6%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL L +E +F E+ L L L ++YRDLK N+L+D +GH ++ D
Sbjct 143 GGDLFTRLSKEVMFTEEDVKFYLAELALALDHLHSLGIIYRDLKPENILLDEEGHIKITD 202
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL+K+ KR +SFCG+ EY++PE++ GH S D ++ G L++E+L G PF
Sbjct 203 FGLSKEAIDHDKRAYSFCGTIEYMAPEVVNRRGHTQSADWWSFGVLMFEMLTGSLPFQGK 262
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWFAG 179
+R I++ +L P S EA+SL+ L P R+GA + GVEE+ +H +F
Sbjct 263 DRKETMALILKAKLGMPQFL--SGEAQSLLRALFKRNPCNRLGAGIDGVEEIKRHPFFVT 320
Query 180 VSWEAA---------KKLTGQLKRTFSFCGSPEYLS 206
+ W K G+ + TF F PE+ +
Sbjct 321 IDWNTLYRKEIKPPFKPAVGRPEDTFHF--DPEFTA 354
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 57/200 (28%), Positives = 100/200 (50%), Gaps = 13/200 (6%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLI-DLDGH---C 56
GG+L + + SE A V + + L Q VV+RDLK +N+L D G
Sbjct 491 GGELLDRILRQRYFSEREASDVLCTITKTMDYLHSQGVVHRDLKPSNILYRDESGSPESI 550
Query 57 RLADFGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPP 116
R+ DFG AK+L + C + +++PE+L G+D + D+++LG L+Y +L GF P
Sbjct 551 RVCDFGFAKQLRAGNGLLMTPCYTANFVAPEVLKRQGYDAACDIWSLGILLYTMLAGFTP 610
Query 117 FFAANRNV---MYMKIMQGQLSFPASC--QASAEARSLVSRLLSLEPLMRVGALGGVEEV 171
F + + +I G+ + S A+ +VS++L ++P R+ A+ +V
Sbjct 611 FANGPDDTPEEILARIGSGKYALSGGNWDSISDAAKDVVSKMLHVDPHQRLTAM----QV 666
Query 172 MQHSWFAGVSWEAAKKLTGQ 191
++H W + + +L+ Q
Sbjct 667 LKHPWVVNREYLSPNQLSRQ 686
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 185 AKKLTGQLKRTFSFCGSPEYLSPEML 210
+K+ KR +SFCG+ EY++PE++
Sbjct 206 SKEAIDHDKRAYSFCGTIEYMAPEVV 231
> Hs4885563
Length=737
Score = 137 bits (344), Expect = 2e-32, Method: Composition-based stats.
Identities = 69/185 (37%), Positives = 108/185 (58%), Gaps = 5/185 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GGDL ++ ++ E +RF A EV L L Q V+YRDLK N+L+D +GHC+LAD
Sbjct 491 GGDLMFQIQRSRKFDEPRSRFYAAEVTSALMFLHQHGVIYRDLKLDNILLDAEGHCKLAD 550
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FG+ K+ T +FCG+P+Y++PE+L + SVD +ALG L+YE++ G PPF A
Sbjct 551 FGMCKEGILNGVTTTTFCGTPDYIAPEILQELEYGPSVDWWALGVLMYEMMAGQPPFEAD 610
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALG---GVEEVMQHSWF 177
N + ++ I+ + +P S EA S++ ++ P R+G + G + + QH +F
Sbjct 611 NEDDLFESILHDDVLYPV--WLSKEAVSILKAFMTKNPHKRLGCVASQNGEDAIKQHPFF 668
Query 178 AGVSW 182
+ W
Sbjct 669 KEIDW 673
> Hs5032091
Length=431
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 76/211 (36%), Positives = 109/211 (51%), Gaps = 4/211 (1%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L+ + E ARF A E+ L L +VYRDLK N+L+D GH L D
Sbjct 181 GGELFYHLQRERCFLEPRARFYAAEIASALGYLHSLNIVYRDLKPENILLDSQGHIVLTD 240
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K+ T +FCG+PEYL+PE+L +D +VD + LG ++YE+L G PPF++
Sbjct 241 FGLCKENIEHNSTTSTFCGTPEYLAPEVLHKQPYDRTVDWWCLGAVLYEMLYGLPPFYSR 300
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGALGGVEEVMQHSWFAGV 180
N MY I+ L + S AR L+ LL + R+GA E+ H +F+ +
Sbjct 301 NTAEMYDNILNKPLQLKPNITNS--ARHLLEGLLQKDRTKRLGAKDDFMEIKSHVFFSLI 358
Query 181 SWE--AAKKLTGQLKRTFSFCGSPEYLSPEM 209
+W+ KK+T S + PE
Sbjct 359 NWDDLINKKITPPFNPNVSGPNELRHFDPEF 389
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 11/16 (68%), Positives = 15/16 (93%), Gaps = 0/16 (0%)
Query 195 TFSFCGSPEYLSPEML 210
T +FCG+PEYL+PE+L
Sbjct 254 TSTFCGTPEYLAPEVL 269
> Hs4885549
Length=479
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 73/211 (34%), Positives = 118/211 (55%), Gaps = 5/211 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG+L L + SE RF E++ L L ++VYRDLK N+++D DGH ++ D
Sbjct 230 GGELFFHLSRERVFSEDRTRFYGAEIVSALDYLHSGKIVYRDLKLENLMLDKDGHIKITD 289
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGL K+ +FCG+PEYL+PE+L + + +VD + LG ++YE++ G PF+
Sbjct 290 FGLCKEGITDAATMKTFCGTPEYLAPEVLEDNDYGRAVDWWGLGVVMYEMMCGRLPFYNQ 349
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRV-GALGGVEEVMQHSWFAG 179
+ ++ I+ + FP + S++A+SL+S LL +P R+ G +E+M+HS+F+G
Sbjct 350 DHEKLFELILMEDIKFPRTL--SSDAKSLLSGLLIKDPNKRLGGGPDDAKEIMRHSFFSG 407
Query 180 VSWEAA--KKLTGQLKRTFSFCGSPEYLSPE 208
V+W+ KKL K + Y E
Sbjct 408 VNWQDVYDKKLVPPFKPQVTSETDTRYFDEE 438
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 10/14 (71%), Positives = 14/14 (100%), Gaps = 0/14 (0%)
Query 197 SFCGSPEYLSPEML 210
+FCG+PEYL+PE+L
Sbjct 305 TFCGTPEYLAPEVL 318
> At3g08720
Length=471
Score = 136 bits (342), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 68/184 (36%), Positives = 109/184 (59%), Gaps = 5/184 (2%)
Query 1 GGDLAALLRCHKRLSEAAARFVAVEVMLGLQQLQQQQVVYRDLKAANVLIDLDGHCRLAD 60
GG L L E AR E++ + L ++ +++RDLK N+L+D+DGH L D
Sbjct 222 GGHLFFQLYHQGLFREDLARVYTAEIVSAVSHLHEKGIMHRDLKPENILMDVDGHVMLTD 281
Query 61 FGLAKKLTGQLKRTFSFCGSPEYLSPEMLLGSGHDHSVDLYALGCLIYELLVGFPPFFAA 120
FGLAK+ + R+ S CG+ EY++PE++ G GHD + D +++G L+YE+L G PPF +
Sbjct 282 FGLAKEFE-ENTRSNSMCGTTEYMAPEIVRGKGHDKAADWWSVGILLYEMLTGKPPFLGS 340
Query 121 NRNVMYMKIMQGQLSFPASCQASAEARSLVSRLLSLEPLMRVGA-LGGVEEVMQHSWFAG 179
+ + KI++ ++ P S EA +L+ LL EP R+G+ G EE+ +H WF
Sbjct 341 -KGKIQQKIVKDKIKLPQFL--SNEAHALLKGLLQKEPERRLGSGPSGAEEIKKHKWFKA 397
Query 180 VSWE 183
++W+
Sbjct 398 INWK 401
Lambda K H
0.323 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3897491854
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40