bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1868_orf2
Length=110
Score E
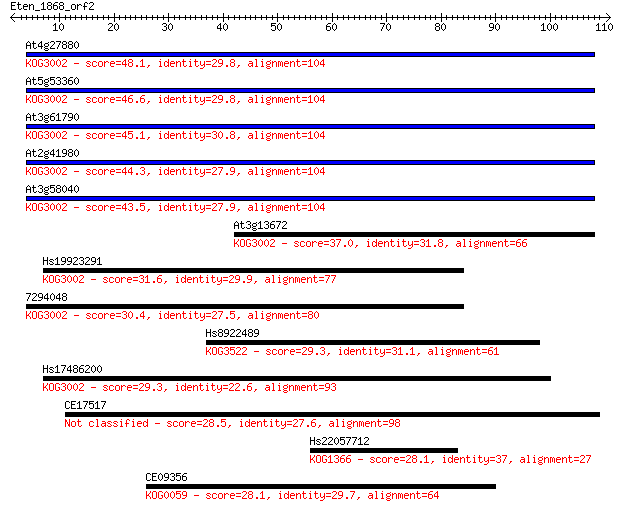
Sequences producing significant alignments: (Bits) Value
At4g27880 48.1 4e-06
At5g53360 46.6 1e-05
At3g61790 45.1 4e-05
At2g41980 44.3 6e-05
At3g58040 43.5 1e-04
At3g13672 37.0 0.009
Hs19923291 31.6 0.35
7294048 30.4 0.80
Hs8922489 29.3 1.9
Hs17486200 29.3 2.0
CE17517 28.5 3.6
Hs22057712 28.1 4.0
CE09356 28.1 4.3
> At4g27880
Length=327
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 51/107 (47%), Gaps = 8/107 (7%)
Query 4 LYHCFGKYFVLRVHRKVEAEAAFYVSVVALHPRHHCSR---YSLQVSGNHRAYSFQGPVW 60
++HCFG+YF L Y++ + +R YSL+V G+ R +++G
Sbjct 207 VFHCFGQYFCLHFEAFQLGMGPVYMAFLRFMGDEEDARSYSYSLEVGGSGRKLTWEGTPR 266
Query 61 SASRGFKELERVKDCLLLPENIALFLSGAKGSEQNLNAISLSVTGEI 107
S +++ D L++ N+ALF SG E + L VTG+I
Sbjct 267 SIRDSHRKVRDSNDGLIIQRNMALFFSGGDRKE-----LKLRVTGKI 308
> At5g53360
Length=305
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 50/107 (46%), Gaps = 8/107 (7%)
Query 4 LYHCFGKYFVLRVHRKVEAEAAFYVSVVALHPRHHCSR---YSLQVSGNHRAYSFQGPVW 60
++ CFG+YF L A Y++ + +R YSL+V G+ R +++G
Sbjct 185 VFQCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDEDDARNYTYSLEVGGSGRKQTWEGTPR 244
Query 61 SASRGFKELERVKDCLLLPENIALFLSGAKGSEQNLNAISLSVTGEI 107
S +++ D L++ N+ALF SG E + L VTG I
Sbjct 245 SVRDSHRKVRDSHDGLIIQRNMALFFSGGDKKE-----LKLRVTGRI 286
> At3g61790
Length=315
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 49/107 (45%), Gaps = 8/107 (7%)
Query 4 LYHCFGKYFVLRVHRKVEAEAAFYVSVVALHPRHHCSR---YSLQVSGNHRAYSFQGPVW 60
++HCFG+YF L A Y++ + +R YSL+V G R ++G
Sbjct 195 VFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDETEARNYNYSLEVGGYGRKLIWEGTPR 254
Query 61 SASRGFKELERVKDCLLLPENIALFLSGAKGSEQNLNAISLSVTGEI 107
S +++ D L++ N+ALF SG E + L VTG I
Sbjct 255 SVRDSHRKVRDSHDGLIIQRNMALFFSGGDRKE-----LKLRVTGRI 296
> At2g41980
Length=305
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 53/107 (49%), Gaps = 8/107 (7%)
Query 4 LYHCFGKYFVLRVHRKVEAEAAFYVSVVALHPRHHCSR---YSLQVSGNHRAYSFQGPVW 60
+++CFG+ F L A Y++ + + ++ YSL+V + R ++QG
Sbjct 200 VFNCFGRQFCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHSRKLTWQGIPR 259
Query 61 SASRGFKELERVKDCLLLPENIALFLSGAKGSEQNLNAISLSVTGEI 107
S +++ +D L++P N+AL+ SG+ E + L VTG I
Sbjct 260 SIRDSHRKVRDSQDGLIIPRNLALYFSGSDKEE-----LKLRVTGRI 301
> At3g58040
Length=308
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 52/107 (48%), Gaps = 8/107 (7%)
Query 4 LYHCFGKYFVLRVHRKVEAEAAFYVSVVALHPRHHCSR---YSLQVSGNHRAYSFQGPVW 60
+++CFG+ F L A Y++ + + ++ YSL+V + R ++QG
Sbjct 203 VFNCFGRQFCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHGRKLTWQGIPR 262
Query 61 SASRGFKELERVKDCLLLPENIALFLSGAKGSEQNLNAISLSVTGEI 107
S +++ +D L++P N+AL+ SG E + L VTG I
Sbjct 263 SIRDSHRKVRDSQDGLIIPRNLALYFSGGDRQE-----LKLRVTGRI 304
> At3g13672
Length=216
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query 42 YSLQVSGNHRAYSFQGPVWSASRGFKELERVKDCLLLPENIALFLSGAKGSEQNLNAISL 101
YSLQV GN R ++QG S K + +D L++ +ALF S + + L
Sbjct 145 YSLQVGGNGRKLTWQGVPRSIRDSHKTVRDSQDGLIITRKLALFFSTDNNTTD--KELKL 202
Query 102 SVTGEI 107
V+G +
Sbjct 203 KVSGRV 208
> Hs19923291
Length=324
Score = 31.6 bits (70), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 34/81 (41%), Gaps = 4/81 (4%)
Query 7 CFGKYF--VLRVHRKVEAEAAFYVSVVALHPRHHCSR--YSLQVSGNHRAYSFQGPVWSA 62
CFG +F VL K E F+ V+ + R Y L+++GN R +++ S
Sbjct 224 CFGHHFMLVLEKQEKYEGHQQFFAIVLLIGTRKQAENFAYRLELNGNRRRLTWEATPRSI 283
Query 63 SRGFKELERVKDCLLLPENIA 83
G DCL+ IA
Sbjct 284 HDGVAAAIMNSDCLVFDTAIA 304
> 7294048
Length=314
Score = 30.4 bits (67), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 35/84 (41%), Gaps = 4/84 (4%)
Query 4 LYHCFGKYF--VLRVHRKVEAEAAFYVSVVALHPRHHCSR--YSLQVSGNHRAYSFQGPV 59
+ CFG +F VL K + F+ V + R Y L+++GN R +++
Sbjct 213 MQSCFGHHFMLVLEKQEKYDGHQQFFAIVQLIGSRKEAENFVYRLELNGNRRRLTWEAMP 272
Query 60 WSASRGFKELERVKDCLLLPENIA 83
S G DCL+ +IA
Sbjct 273 RSIHEGVASAIHNSDCLVFDTSIA 296
> Hs8922489
Length=596
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 4/61 (6%)
Query 37 HHCSRYSLQVSGNHRAYSFQGPVWSASRGFKELERVKDCLLLPENIALFLSGAKGSEQNL 96
H SG + + GP R F+EL+ ++ L + E I L +S G+ QNL
Sbjct 152 QHSGAKKASASGQAQNKVYLGP----PRLFQELQDLQKDLAVVEQITLLISTLHGTYQNL 207
Query 97 N 97
N
Sbjct 208 N 208
> Hs17486200
Length=378
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 21/97 (21%), Positives = 41/97 (42%), Gaps = 6/97 (6%)
Query 7 CFGKYFVLRVHRKVEAEAA--FYVSVVALHPRHHCSR--YSLQVSGNHRAYSFQGPVWSA 62
CFG F+L + ++ + F+ V L Y L++ GN R +++
Sbjct 280 CFGVRFMLVLQKQEDHNGGQQFFAVVQLLGASKEAENFAYQLELKGNRRRLTWEATPLPI 339
Query 63 SRGFKELERVKDCLLLPENIALFLSGAKGSEQNLNAI 99
+ + +DCL+ N AL A+ + ++N +
Sbjct 340 HEDIAKAIKNRDCLIFDANTALLF--AENDDLSINVV 374
> CE17517
Length=684
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 27/110 (24%), Positives = 47/110 (42%), Gaps = 12/110 (10%)
Query 11 YFVLRVHRKVEAEAAFYV---SVVALHPRHHCSRYSLQVSGNH-RAYSFQGPVWSASRGF 66
+F + + V YV LH +H+C + +L GN A PV ++S +
Sbjct 280 HFFITAYNNVSKRIGHYVFNQKFGQLHKQHYCPKCTLFDKGNRVNAEQVIMPVVTSSGEW 339
Query 67 -------KELERVKDCLLLPENIALFLSGAK-GSEQNLNAISLSVTGEIL 108
KE +D L PE L + G G+ ++N I+ S+ +++
Sbjct 340 PIVLCTDKEGRLCRDSFLKPEGTFLKVPGQTLGATMDINEINTSLKADVV 389
> Hs22057712
Length=1857
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 56 QGPVWSASRGFKELERVKDCLLLPENI 82
+G +W ++ FK LER+K + + EN+
Sbjct 1327 KGSIWLSALTFKTLERMKKFVFIDENV 1353
> CE09356
Length=1447
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 25/64 (39%), Gaps = 0/64 (0%)
Query 26 FYVSVVALHPRHHCSRYSLQVSGNHRAYSFQGPVWSASRGFKELERVKDCLLLPENIALF 85
Y V +HPRHHC+ L V R Y + + V D L L +LF
Sbjct 972 LYDMVGGIHPRHHCNNAHLPVLPCLRLYRRRRNILRLPPSILRARNVDDSLRLCIPKSLF 1031
Query 86 LSGA 89
G+
Sbjct 1032 CGGS 1035
Lambda K H
0.322 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40