bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1876_orf1
Length=137
Score E
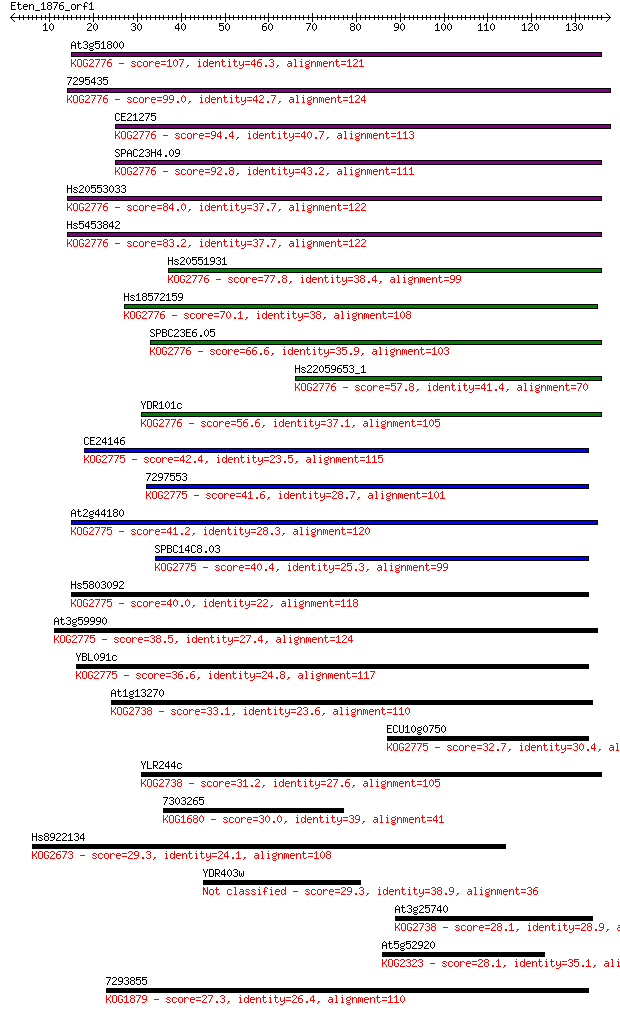
Sequences producing significant alignments: (Bits) Value
At3g51800 107 5e-24
7295435 99.0 2e-21
CE21275 94.4 6e-20
SPAC23H4.09 92.8 2e-19
Hs20553033 84.0 9e-17
Hs5453842 83.2 1e-16
Hs20551931 77.8 6e-15
Hs18572159 70.1 1e-12
SPBC23E6.05 66.6 1e-11
Hs22059653_1 57.8 6e-09
YDR101c 56.6 1e-08
CE24146 42.4 3e-04
7297553 41.6 4e-04
At2g44180 41.2 6e-04
SPBC14C8.03 40.4 0.001
Hs5803092 40.0 0.001
At3g59990 38.5 0.004
YBL091c 36.6 0.014
At1g13270 33.1 0.18
ECU10g0750 32.7 0.20
YLR244c 31.2 0.53
7303265 30.0 1.3
Hs8922134 29.3 2.2
YDR403w 29.3 2.4
At3g25740 28.1 5.2
At5g52920 28.1 5.6
7293855 27.3 9.0
> At3g51800
Length=392
Score = 107 bits (268), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 56/122 (45%), Positives = 80/122 (65%), Gaps = 4/122 (3%)
Query 15 SEGEREGPQ-ELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEA 73
S+ ER+ + L+SP+ V K K+AAEI N AL+ VLA P A + ++C GD F+ ++
Sbjct 3 SDDERDEKELSLTSPEVVTKYKSAAEIVNKALQVVLAECKPKAKIVDICEKGDSFIKEQT 62
Query 74 AKVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGF 133
A +Y K KKIE+G+A PTC+SVN +FSP S +L +GD+VK+ +G HIDGF
Sbjct 63 ASMY--KNSKKKIERGVAFPTCISVNNTVGHFSPLASDES-VLEDGDMVKIDMGCHIDGF 119
Query 134 VA 135
+A
Sbjct 120 IA 121
> 7295435
Length=391
Score = 99.0 bits (245), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 53/124 (42%), Positives = 74/124 (59%), Gaps = 3/124 (2%)
Query 14 LSEGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEA 73
+++ E+E + ++ V K K A EI N L+ V+ V A V E+C GD + +E
Sbjct 1 MADVEKEPEKTIAEDLVVTKYKLAGEIVNKTLKAVIGLCVVDASVREICTQGDNQLTEET 60
Query 74 AKVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGF 133
KVY+ K K ++KGIA PTC+SVN +FSP A L+ GD+VK+ LGAHIDGF
Sbjct 61 GKVYK---KEKDLKKGIAFPTCLSVNNCVCHFSPAKNDADYTLKAGDVVKIDLGAHIDGF 117
Query 134 VAAA 137
+A A
Sbjct 118 IAVA 121
> CE21275
Length=391
Score = 94.4 bits (233), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 70/113 (61%), Gaps = 3/113 (2%)
Query 25 LSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVYQRKAKAK 84
L++ V K + AAEI NA L++VLA + GA +LC GD + ++ K+Y+ K K
Sbjct 23 LANDAVVTKYQVAAEITNAVLKEVLANIKEGAIAGDLCDLGDKLILEKTGKLYK---KEK 79
Query 85 KIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGFVAAA 137
KGIA+PTC+S++ +++P A ++L+ G +VKV LG HIDG +A A
Sbjct 80 NFTKGIAMPTCISIDNCICHYTPLKSEAPVVLKNGQVVKVDLGTHIDGLIATA 132
> SPAC23H4.09
Length=381
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 48/113 (42%), Positives = 71/113 (62%), Gaps = 6/113 (5%)
Query 25 LSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVYQRKAKAK 84
LS+P+ V K K A E++ +++V+ PGA ++++C+ GD + + KVY + K
Sbjct 16 LSNPETVNKYKIAGEVSQNVIKKVVELCQPGAKIYDICVRGDELLNEAIKKVY----RTK 71
Query 85 KIEKGIAVPTCVSVNEMFANFSP--CDRSASLLLREGDLVKVHLGAHIDGFVA 135
KGIA PT VS N+M A+ SP D A+L L+ GD+VK+ LGAHIDGF +
Sbjct 72 DAYKGIAFPTAVSPNDMAAHLSPLKSDPEANLALKSGDVVKILLGAHIDGFAS 124
> Hs20553033
Length=210
Score = 84.0 bits (206), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 46/122 (37%), Positives = 69/122 (56%), Gaps = 3/122 (2%)
Query 14 LSEGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEA 73
+S + + Q ++ V K K +IAN LR ++ G V LC GD + +E
Sbjct 1 MSGEDEQQEQTIAEDLVVTKYKMGGDIANRVLRSLVEASSSGVSVLSLCEKGDAMIMEET 60
Query 74 AKVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGF 133
K+++ K K+++KGIA PT +SVN +FSP +L+EGDLVK+ LG H+DGF
Sbjct 61 GKIFK---KEKEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGF 117
Query 134 VA 135
+A
Sbjct 118 IA 119
> Hs5453842
Length=394
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 46/122 (37%), Positives = 69/122 (56%), Gaps = 3/122 (2%)
Query 14 LSEGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEA 73
+S + + Q ++ V K K +IAN LR ++ G V LC GD + +E
Sbjct 1 MSGEDEQQEQTIAEDLVVTKYKMGGDIANRVLRSLVEASSSGVSVLSLCEKGDAMIMEET 60
Query 74 AKVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGF 133
K+++ K K+++KGIA PT +SVN +FSP +L+EGDLVK+ LG H+DGF
Sbjct 61 GKIFK---KEKEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGF 117
Query 134 VA 135
+A
Sbjct 118 IA 119
> Hs20551931
Length=307
Score = 77.8 bits (190), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 59/99 (59%), Gaps = 3/99 (3%)
Query 37 AAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVYQRKAKAKKIEKGIAVPTCV 96
+IA+ LR ++ G V LC D + +E K+++ K K+++KGIA PT +
Sbjct 2 GGDIAHRVLRSLVEASSSGVLVLSLCEKSDAVIMEETGKIFK---KEKEMKKGIAFPTSI 58
Query 97 SVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGFVA 135
SVN +FSP + +L+EGDLVK+ LG ++DGF+A
Sbjct 59 SVNNCVCHFSPLESGQDYILKEGDLVKIDLGVYVDGFIA 97
> Hs18572159
Length=910
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 41/109 (37%), Positives = 61/109 (55%), Gaps = 4/109 (3%)
Query 27 SPQAVEKLKA-AAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVYQRKAKAKK 85
SP+ L +IAN LR ++ G V LC GD + +E K+++ K K+
Sbjct 421 SPRPPHSLTGWGGDIANGVLRSLVDASSSGVSVLSLCEKGDAVIMQETGKIFK---KEKE 477
Query 86 IEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGFV 134
++K IA PT + VN +FSP R +L+EGDLVK+ LG H++GF+
Sbjct 478 MKKRIAFPTSILVNNCVCHFSPLKRDQDYILKEGDLVKIDLGVHVNGFI 526
> SPBC23E6.05
Length=417
Score = 66.6 bits (161), Expect = 1e-11, Method: Composition-based stats.
Identities = 37/109 (33%), Positives = 58/109 (53%), Gaps = 10/109 (9%)
Query 33 KLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVYQRKAKAKKIEKGIAV 92
K + A + + A QV +R VPGA E+ +GD + + + +Y K+++ EKGIA
Sbjct 20 KYRDAGALVSKAFHQVASRCVPGASTREISSYGDNLLHEYKSSIY----KSQRFEKGIAE 75
Query 93 PTCVSVNEMFANFSPCDRSA------SLLLREGDLVKVHLGAHIDGFVA 135
PT + VN N++P S S L+ GD+ K+ +G H DG+ A
Sbjct 76 PTSICVNNCAYNYAPGPESVIAGNDNSYHLQVGDVTKISMGLHFDGYTA 124
> Hs22059653_1
Length=215
Score = 57.8 bits (138), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 44/73 (60%), Gaps = 6/73 (8%)
Query 66 DLFVAK---EAAKVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLV 122
DL + K E K+++ K K+++K IA PT + VN +FSP +L+EG+LV
Sbjct 15 DLVMTKYQMETGKIFK---KVKEMKKSIAFPTSILVNNCVGHFSPLKSDKDCILKEGNLV 71
Query 123 KVHLGAHIDGFVA 135
K+ LG +DGF+A
Sbjct 72 KIDLGVQVDGFIA 84
> YDR101c
Length=593
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 39/132 (29%), Positives = 61/132 (46%), Gaps = 31/132 (23%)
Query 31 VEKLKAAAEIANAALRQVLARVVPG---------ADVFELCLFGDLFVAKEAAKVYQRKA 81
+ K + A +IA AL+ V + + V ELCL D F+ + Y+ K
Sbjct 25 LNKYRTAGQIAQTALKYVTSLINDSYHSKTTQRQLTVPELCLLTDSFILTRLEQYYKNKV 84
Query 82 KAKKIEKGIAVPTCVSVNEMFANFSP-CDRSASLL-----------------LREGDLVK 123
E+GIA+PT + ++++ + P D + +LL LR GDLVK
Sbjct 85 N----ERGIAIPTTIDIDQISGGWCPEIDDTQNLLNWNKGKDSTFASSVTGTLRPGDLVK 140
Query 124 VHLGAHIDGFVA 135
+ LG HIDG+ +
Sbjct 141 ITLGVHIDGYTS 152
> CE24146
Length=444
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 27/115 (23%), Positives = 55/115 (47%), Gaps = 9/115 (7%)
Query 18 EREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVY 77
E + ++S + + + +AE + V + + PG + E+C +
Sbjct 117 EEKKALDISYEEVWQDYRRSAEAHRQVRKYVKSWIKPGMTMIEIC--------ERLETTS 168
Query 78 QRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDG 132
+R K + +E G+A PT S+N A+++P + + +L+ GD+ K+ G H+ G
Sbjct 169 RRLIKEQGLEAGLAFPTGCSLNHCAAHYTP-NAGDTTVLQYGDVCKIDYGIHVRG 222
> 7297553
Length=448
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 29/102 (28%), Positives = 51/102 (50%), Gaps = 11/102 (10%)
Query 32 EKLKAAAEIANAALRQVLARVV-PGADVFELCLFGDLFVAKEAAKVYQRKAKAKKIEKGI 90
++L+ AAE A+ RQ + R + PG + ++C +E +R +E G+
Sbjct 137 QELRQAAE-AHRQTRQYMQRYIKPGMTMIQIC--------EELENTARRLIGENGLEAGL 187
Query 91 AVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDG 132
A PT S+N A+++P + +L+ D+ K+ G HI G
Sbjct 188 AFPTGCSLNHCAAHYTP-NAGDPTVLQYDDVCKIDFGTHIKG 228
> At2g44180
Length=431
Score = 41.2 bits (95), Expect = 6e-04, Method: Composition-based stats.
Identities = 34/121 (28%), Positives = 61/121 (50%), Gaps = 13/121 (10%)
Query 15 SEGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAA 74
SE +RE + L P L+ AAE+ + + + + PG + +LC E
Sbjct 105 SEEKRE-MERLQKP-IYNSLRQAAEVHRQVRKYMRSILKPGMLMIDLC---------ETL 153
Query 75 KVYQRKAKAKK-IEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGF 133
+ RK ++ ++ GIA PT S+N + A+++P + +L+ D++K+ G HIDG
Sbjct 154 ENTVRKLISENGLQAGIAFPTGCSLNNVAAHWTP-NSGDKTVLQYDDVMKLDFGTHIDGH 212
Query 134 V 134
+
Sbjct 213 I 213
> SPBC14C8.03
Length=426
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 25/99 (25%), Positives = 49/99 (49%), Gaps = 9/99 (9%)
Query 34 LKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVYQRKAKAKKIEKGIAVP 93
L+ AAE+ A + + + PG + + V + + ++ GI P
Sbjct 117 LRRAAEVHRQARQYAQSVIKPGMSMMD--------VVNTIENTTRALVEEDGLKSGIGFP 168
Query 94 TCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDG 132
T VS+N A+++P + + +L+E D++KV +G H++G
Sbjct 169 TGVSLNHCAAHYTP-NAGDTTILKEKDVMKVDIGVHVNG 206
> Hs5803092
Length=478
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 26/118 (22%), Positives = 54/118 (45%), Gaps = 9/118 (7%)
Query 15 SEGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAA 74
+ E + + +S + + AAE + V++ + PG + E+C ++
Sbjct 150 TTSEEKKALDQASEEIWNDFREAAEAHRQVRKYVMSWIKPGMTMIEIC--------EKLE 201
Query 75 KVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDG 132
++ K + G+A PT S+N A+++P + + +L+ D+ K+ G HI G
Sbjct 202 DCSRKLIKENGLNAGLAFPTGCSLNNCAAHYTP-NAGDTTVLQYDDICKIDFGTHISG 258
> At3g59990
Length=435
Score = 38.5 bits (88), Expect = 0.004, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 61/125 (48%), Gaps = 13/125 (10%)
Query 11 DHELSEGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVA 70
D SE +RE + P ++ AAE+ + V + V PG + ++C
Sbjct 105 DETTSEEKRE-LERFEKP-IYNSVRRAAEVHRQVRKYVRSIVKPGMLMTDIC-------- 154
Query 71 KEAAKVYQRKAKAKK-IEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAH 129
E + RK ++ ++ GIA PT S+N + A+++P + +L+ D++K+ G H
Sbjct 155 -ETLENTVRKLISENGLQAGIAFPTGCSLNWVAAHWTP-NSGDKTVLQYDDVMKLDFGTH 212
Query 130 IDGFV 134
IDG +
Sbjct 213 IDGHI 217
> YBL091c
Length=421
Score = 36.6 bits (83), Expect = 0.014, Method: Composition-based stats.
Identities = 29/119 (24%), Positives = 53/119 (44%), Gaps = 6/119 (5%)
Query 16 EGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLF--VAKEA 73
E R ++L + ++ AEI R + R+VPG + ++ D+ ++
Sbjct 87 EESRYLKRDLERAEHWNDVRKGAEIHRRVRRAIKDRIVPGMKLMDI---ADMIENTTRKY 143
Query 74 AKVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDG 132
A +GI PT +S+N A+F+P ++L E D++KV G ++G
Sbjct 144 TGAENLLAMEDPKSQGIGFPTGLSLNHCAAHFTPNAGDKTVLKYE-DVMKVDYGVQVNG 201
> At1g13270
Length=369
Score = 33.1 bits (74), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 26/111 (23%), Positives = 46/111 (41%), Gaps = 12/111 (10%)
Query 24 ELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFEL-CLFGDLFVAKEAAKVYQRKAK 82
++ P+ + K++AA E+A L V P E+ D+ + A Y
Sbjct 125 QIPGPEGIAKMRAACELAARVLNYAGTLVKPSVTTNEIDKAVHDMIIE---AGAYPSPLG 181
Query 83 AKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGF 133
K + C SVNE + P R L+ GD++ + + ++DG+
Sbjct 182 YGGFPKSV----CTSVNECMCHGIPDSRQ----LQSGDIINIDVTVYLDGY 224
> ECU10g0750
Length=358
Score = 32.7 bits (73), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 87 EKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDG 132
GI P +S+N A+++ ++L+E D++K+ G H DG
Sbjct 92 NNGIGFPAGMSMNSCAAHYTVNPGEQDIVLKEDDVLKIDFGTHSDG 137
> YLR244c
Length=387
Score = 31.2 bits (69), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 46/107 (42%), Gaps = 14/107 (13%)
Query 31 VEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAK--VYQRKAKAKKIEK 88
++K++ A + L A V PG EL D V E K Y K
Sbjct 135 IKKIRKACMLGREVLDIAAAHVRPGITTDEL----DEIVHNETIKRGAYPSPLNYYNFPK 190
Query 89 GIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGFVA 135
+ C SVNE+ + P D++ +L+EGD+V + + + G+ A
Sbjct 191 SL----CTSVNEVICHGVP-DKT---VLKEGDIVNLDVSLYYQGYHA 229
> 7303265
Length=295
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 36 AAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKV 76
A I A Q L RVV + E+CL G++ A+EA K+
Sbjct 171 ALGTIPGAGGTQRLTRVVGKSKAMEMCLTGNMIGAQEAEKL 211
> Hs8922134
Length=469
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 26/108 (24%), Positives = 39/108 (36%), Gaps = 15/108 (13%)
Query 6 CSKMSDHELSEGEREGPQELSSPQAVEKLKAAAEIANAALRQVLARVVPGADVFELCLFG 65
C D + EG+ Q L P+ E +E +A+ P ++V LC
Sbjct 321 CPNELDLPVPEGKTSEKQTLDEPEVPEIFTKKSEAGHAS--------SPDSEVTSLC--- 369
Query 66 DLFVAKEAAKVYQRKAKAKKIEKGIAVPTCVSVNEMFANFSPCDRSAS 113
KE A++ + ++ G VP C N P D S S
Sbjct 370 ----QKEKAELAPVNTEGALLDNGSVVPNCDISNGGSQKLFPADTSPS 413
> YDR403w
Length=536
Score = 29.3 bits (64), Expect = 2.4, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 1/37 (2%)
Query 45 LRQVLARV-VPGADVFELCLFGDLFVAKEAAKVYQRK 80
L ++ RV +PG D C DLF + A+KV+ K
Sbjct 291 LHELYKRVAIPGVDAIGFCGLNDLFFSGAASKVFDPK 327
> At3g25740
Length=344
Score = 28.1 bits (61), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query 89 GIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDGF 133
G C SVNE + P R L+ GD++ + + ++DG+
Sbjct 157 GFPKSVCTSVNECMFHGIPDSRP----LQNGDIINIDVAVYLDGY 197
> At5g52920
Length=579
Score = 28.1 bits (61), Expect = 5.6, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 24/37 (64%), Gaps = 2/37 (5%)
Query 86 IEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLV 122
IE+G++ P+CVSVN + +F + +LL +G ++
Sbjct 202 IERGVSTPSCVSVN--YDDFVNDVEAGDMLLVDGGMM 236
> 7293855
Length=1548
Score = 27.3 bits (59), Expect = 9.0, Method: Composition-based stats.
Identities = 29/112 (25%), Positives = 45/112 (40%), Gaps = 2/112 (1%)
Query 23 QELSSPQ--AVEKLKAAAEIANAALRQVLARVVPGADVFELCLFGDLFVAKEAAKVYQRK 80
Q+L P+ VE ++A ++ N L + V D+ L L G F A A +
Sbjct 1052 QQLQVPENWLVEAVRAVYDLDNIKLTDIGGPVHSEFDLEYLLLEGHCFDAASGAPPRGLQ 1111
Query 81 AKAKKIEKGIAVPTCVSVNEMFANFSPCDRSASLLLREGDLVKVHLGAHIDG 132
+ V T V N + + SL LREG ++ +HI+G
Sbjct 1112 LVLGTQSQPTLVDTIVMANLGYFQLKANPGAWSLRLREGKSADIYAISHIEG 1163
Lambda K H
0.319 0.132 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40