bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1889_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
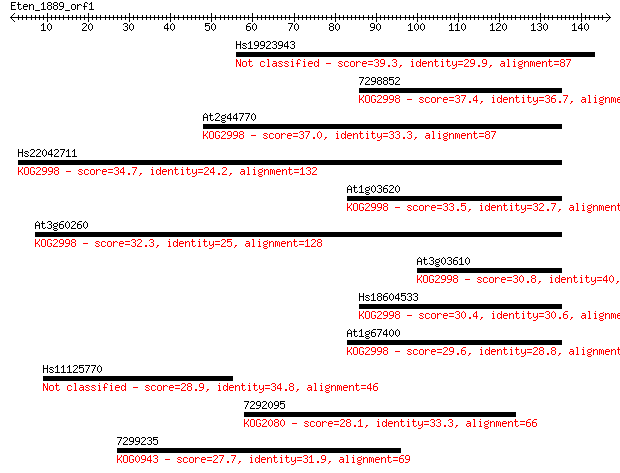
Hs19923943 39.3 0.002
7298852 37.4 0.011
At2g44770 37.0 0.012
Hs22042711 34.7 0.058
At1g03620 33.5 0.15
At3g60260 32.3 0.29
At3g03610 30.8 1.00
Hs18604533 30.4 1.3
At1g67400 29.6 2.1
Hs11125770 28.9 3.8
7292095 28.1 5.5
7299235 27.7 7.8
> Hs19923943
Length=475
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 43/87 (49%), Gaps = 1/87 (1%)
Query 56 SGSSCMRLFAFLSNMFDGEMTLDEKLVLQILRRLAATPYDKENADHEQLLRRFFLLCFPE 115
S S+ + L + + D LDEKL+L+ +R AA P D+ + E+ + C E
Sbjct 193 SSSTNVDLLVKVGEVVDKLFDLDEKLMLEWVRNGAAQPLDQPQEESEEQPVFRLVPCILE 252
Query 116 ATLEPAKEEDSRWKEVGFQVLIYLSTV 142
A + + E+ W +V +L L+TV
Sbjct 253 AA-KQVRSENPEWLDVYMHILQLLTTV 278
> 7298852
Length=316
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 86 LRRLAATPYDKENADHEQLLRRFFLLCFPEATLEPAKEEDSRWKEVGFQ 134
+ +L A YD +N DHEQ L R + L P+ L +W+++GFQ
Sbjct 126 VEQLRAEKYDSDNLDHEQKLLRLWQLLMPDTPL--TGRVTKQWQDIGFQ 172
> At2g44770
Length=250
Score = 37.0 bits (84), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 7/90 (7%)
Query 48 PPSGGIVGSGSSCM---RLFAFLSNMFDGEMTLDEKLVLQILRRLAATPYDKENADHEQL 104
P S +G G SC+ R + ++ FD +T ++ LQ L+ YD H++
Sbjct 33 PGSAAWLGRGLSCVCAQRRDSDANSTFD--LTPAQEECLQSLQNRIDVAYDSTIPLHQEA 90
Query 105 LRRFFLLCFPEATLEPAKEEDSRWKEVGFQ 134
LR + L FPE L E +WKE+G+Q
Sbjct 91 LRELWKLSFPEEELHGLISE--QWKEMGWQ 118
> Hs22042711
Length=293
Score = 34.7 bits (78), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 32/135 (23%), Positives = 59/135 (43%), Gaps = 16/135 (11%)
Query 3 SSLAAQPASRIQNYFSSGQPLRVLSTASLMRNTGLHHRGG---RERGPPPSGGIVGSGSS 59
+ + AQ RI+N + + +VL A+ + + + +E+ P S
Sbjct 37 TYVGAQRTHRIENSLTYSKN-KVLQKATHVVQSEVDKYVDDIMKEKNINPEKD--ASFKI 93
Query 60 CMRLFAFLSNMFDGEMTLDEKLVLQILRRLAATPYDKENADHEQLLRRFFLLCFPEATLE 119
CM++ ++T ++L L + + PYD +N HE+LL + + L P L
Sbjct 94 CMKMCLL-------QITGYKQLYLDV-ESVRKRPYDSDNLQHEELLMKLWNLLMPTKKLN 145
Query 120 PAKEEDSRWKEVGFQ 134
+ +W E+GFQ
Sbjct 146 ARISK--QWAEIGFQ 158
> At1g03620
Length=265
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query 83 LQILRRLAATPYDKENADHEQLLRRFFLLCFPEATLEPAKEEDSRWKEVGFQ 134
LQ L+ P+D+ DH++ L+ + + FP L E +WKE+G+Q
Sbjct 67 LQRLQDRMVVPFDETRPDHQESLKALWNVAFPNVHLTGLVTE--QWKEMGWQ 116
> At3g60260
Length=250
Score = 32.3 bits (72), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 46/128 (35%), Gaps = 24/128 (18%)
Query 7 AQPASRIQNYFSSGQPLRVLSTASLMRNTGLHHRGGRERGPPPSGGIVGSGSSCMRLFAF 66
+Q R Y SS + S A L R GR+ P PS + + C
Sbjct 15 SQGLERGSVYHSSSAEVVAGSAAWLGRGLSCVCVQGRDGDPRPSFDLTPAQEEC------ 68
Query 67 LSNMFDGEMTLDEKLVLQILRRLAATPYDKENADHEQLLRRFFLLCFPEATLEPAKEEDS 126
LQ L+ YD H++ L+ + L FPE L +
Sbjct 69 ----------------LQRLQSRIDVAYDSSIPQHQEALKDLWKLAFPEEELHGIVSD-- 110
Query 127 RWKEVGFQ 134
+WKE+G+Q
Sbjct 111 QWKEMGWQ 118
> At3g03610
Length=182
Score = 30.8 bits (68), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 2/35 (5%)
Query 100 DHEQLLRRFFLLCFPEATLEPAKEEDSRWKEVGFQ 134
+H+ LR+ + L +P+ L P K E WKE+G+Q
Sbjct 2 EHQDALRQLWRLAYPQRELPPLKSE--LWKEMGWQ 34
> Hs18604533
Length=334
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 86 LRRLAATPYDKENADHEQLLRRFFLLCFPEATLEPAKEEDSRWKEVGFQ 134
+ +L YD +N HE++L + + P LE + +W E+GFQ
Sbjct 119 VEKLRREAYDSDNPQHEEMLLKLWKFLKPNTPLESRISK--QWCEIGFQ 165
> At1g67400
Length=294
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 26/52 (50%), Gaps = 2/52 (3%)
Query 83 LQILRRLAATPYDKENADHEQLLRRFFLLCFPEATLEPAKEEDSRWKEVGFQ 134
L+ LR+ YD DH+ LR + +P+ L+ + +WK +G+Q
Sbjct 85 LKRLRKRMKNYYDASRPDHQDALRALWSATYPDEKLQDLISD--QWKNMGWQ 134
> Hs11125770
Length=320
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 21/46 (45%), Gaps = 3/46 (6%)
Query 9 PASRIQNYFSSGQPLRVLSTASLMRNTGLHHRGGRERGPPPSGGIV 54
P +++ Y G PLRV + R +H GR GP P G V
Sbjct 2 PMYQVKPYHGGGAPLRVELPTCMYRLPNVH---GRSYGPAPGAGHV 44
> 7292095
Length=1449
Score = 28.1 bits (61), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 35/79 (44%), Gaps = 13/79 (16%)
Query 58 SSCMRLFAFLSNMFDGE-------------MTLDEKLVLQILRRLAATPYDKENADHEQL 104
SS RL+ L + +G ++L E L+ ++++ +A T E + E
Sbjct 1352 SSIWRLYVQLMDEINGTALGKDGKFQLLICLSLREHLLTRLIKPMALTKVTHEMYEEESF 1411
Query 105 LRRFFLLCFPEATLEPAKE 123
LRR LL F LEP +
Sbjct 1412 LRRRNLLTFLIQILEPLDD 1430
> 7299235
Length=2165
Score = 27.7 bits (60), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 33/75 (44%), Gaps = 6/75 (8%)
Query 27 STASLMRNTGLHHRGGRERGPPPSGGIVGSGSSCMRLFAFLSNMFDGEMTLDEKLVLQ-- 84
S A L+R TG + G+ G +G I+G +S L + E+ ++VLQ
Sbjct 108 SRARLLRATGRSNSTGQGSGSRSTGVIIGGSTSSRPLVTVPATYVPEELISQAEVVLQGK 167
Query 85 ----ILRRLAATPYD 95
I+R L T D
Sbjct 168 SRNLIIRELQRTNLD 182
Lambda K H
0.320 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40