bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1909_orf1
Length=175
Score E
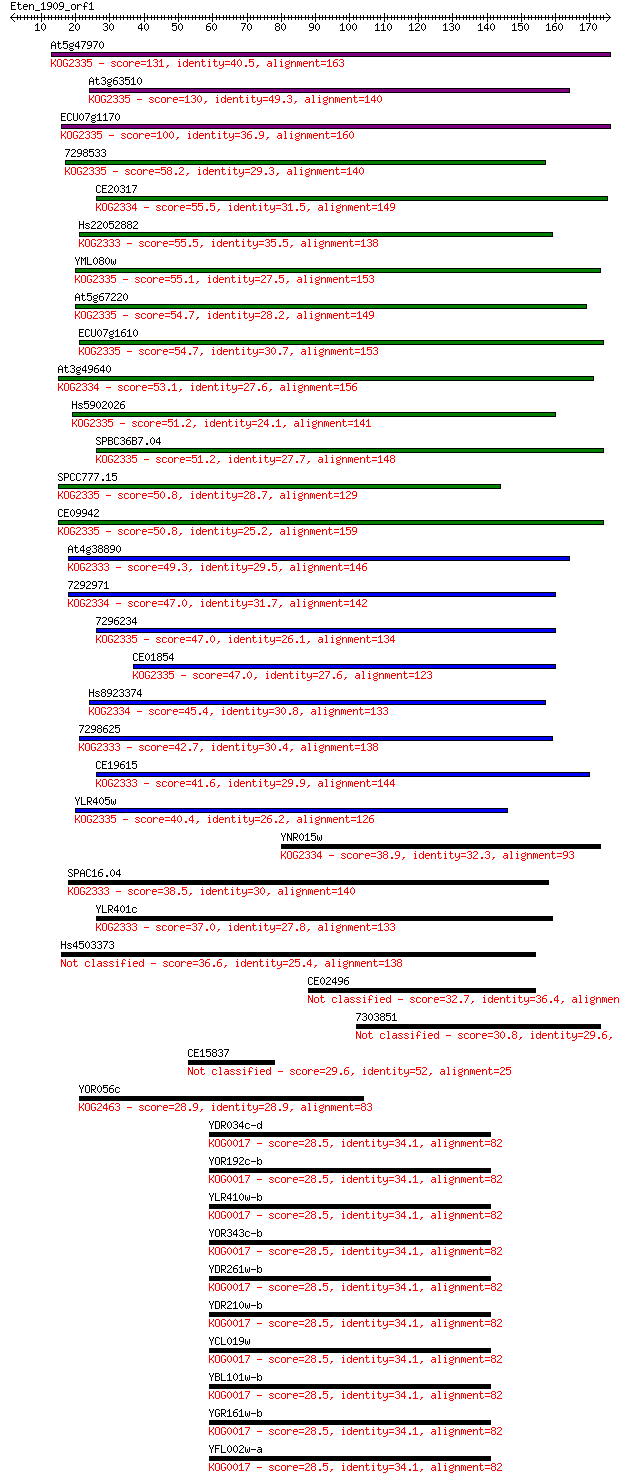
Sequences producing significant alignments: (Bits) Value
At5g47970 131 9e-31
At3g63510 130 1e-30
ECU07g1170 100 1e-21
7298533 58.2 9e-09
CE20317 55.5 5e-08
Hs22052882 55.5 6e-08
YML080w 55.1 7e-08
At5g67220 54.7 8e-08
ECU07g1610 54.7 9e-08
At3g49640 53.1 2e-07
Hs5902026 51.2 1e-06
SPBC36B7.04 51.2 1e-06
SPCC777.15 50.8 1e-06
CE09942 50.8 1e-06
At4g38890 49.3 3e-06
7292971 47.0 2e-05
7296234 47.0 2e-05
CE01854 47.0 2e-05
Hs8923374 45.4 6e-05
7298625 42.7 4e-04
CE19615 41.6 7e-04
YLR405w 40.4 0.002
YNR015w 38.9 0.006
SPAC16.04 38.5 0.007
YLR401c 37.0 0.019
Hs4503373 36.6 0.024
CE02496 32.7 0.41
7303851 30.8 1.3
CE15837 29.6 3.1
YOR056c 28.9 6.0
YDR034c-d 28.5 7.5
YOR192c-b 28.5 8.0
YLR410w-b 28.5 8.0
YOR343c-b 28.5 8.0
YDR261w-b 28.5 8.0
YDR210w-b 28.5 8.0
YCL019w 28.5 8.0
YBL101w-b 28.5 8.0
YGR161w-b 28.5 8.1
YFL002w-a 28.5 8.1
> At5g47970
Length=387
Score = 131 bits (329), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 66/163 (40%), Positives = 95/163 (58%), Gaps = 0/163 (0%)
Query 13 MGETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLG 72
M +E S L ++APM+ ++ H+R RLIT+ + +YTEM+ TI++ L++ L
Sbjct 1 MTVSEAYSPPLFSIAPMMGWTDNHYRTLARLITKHAWLYTEMLAAETIVYQEDNLDSFLA 60
Query 73 FDDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSP 132
F +HPIV Q+GG N +N+A+A A YDE+N N GCPS +V +G FGA LM P
Sbjct 61 FSPDQHPIVLQIGGRNLENLAKATRLANAYAYDEINFNCGCPSPKVSGRGCFGALLMLDP 120
Query 133 TKVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
V + + + VTVKCR+GVD+ DS F+ VS
Sbjct 121 KFVGEAMSVIAANTNAAVTVKCRIGVDDHDSYNELCDFIHIVS 163
> At3g63510
Length=456
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 69/165 (41%), Positives = 95/165 (57%), Gaps = 26/165 (15%)
Query 24 LAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQ 83
+VAPM+ ++ H+R RLIT+ + +YTEM+ T++H L+ L F +HPIV Q
Sbjct 39 FSVAPMMDWTDNHYRTLARLITKHAWLYTEMIAAETLVHQQTNLDRFLAFSPQQHPIVLQ 98
Query 84 LGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIV--YE 141
LGGSN +N+A+AA ++ GYDE+NLN GCPS +V G FG +LM P KVR ++
Sbjct 99 LGGSNVENLAKAAKLSDAYGYDEINLNCGCPSPKVAGHGCFGVSLMLKP-KVRYFTPPHD 157
Query 142 MKRRV-----------------------QIPVTVKCRLGVDNLDS 163
KR + +PVTVKCR+GVDN DS
Sbjct 158 FKRYLLFLNFTLSFKLVGEAMSAIAANTNVPVTVKCRIGVDNHDS 202
> ECU07g1170
Length=357
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 59/162 (36%), Positives = 92/162 (56%), Gaps = 6/162 (3%)
Query 16 TEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILH-NTHQLENHLG-F 73
+ +S +++APM+ V+ HFR F+RL + ++ ++TEM+V +T++H +L LG +
Sbjct 7 SPMSQEVEISLAPMMDVTTAHFRRFIRLTSEKTVLFTEMIVSNTVIHVPRDKLRERLGEY 66
Query 74 DDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPT 133
DD V Q+GGS+ ++ EA + GY NLN GCPS R V KGSFGA LM +
Sbjct 67 DD---RTVVQIGGSDATSMVEAVRILQGLGYRMFNLNCGCPSSR-VKKGSFGAVLMLNRE 122
Query 134 KVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
V +I+ + +++K R GVD D +F FV +
Sbjct 123 LVAEIINRVYGETGAVLSLKIRTGVDEHDGVDFLDGFVSHIK 164
> 7298533
Length=396
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 41/144 (28%), Positives = 67/144 (46%), Gaps = 9/144 (6%)
Query 17 EISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDV 76
E S + APM+ S FR +RL +T M++ +I ++ +N
Sbjct 18 EAQSDFVRVSAPMVRYSKLEFRRLVRL-NGVQLAFTPMMISDSINNSEKARQNEFSTGAD 76
Query 77 EHPIVCQLGGSNPQNVAEAATWAEL--AGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
+ P++ Q +P E T A+L D ++LN GCP + KG +G ++R P
Sbjct 77 DQPLIAQFAAKDP---TEFVTSAQLIYPYVDGIDLNCGCPQSWAMAKG-YGCGMLRQPEL 132
Query 135 VRDIVYEMKRRV--QIPVTVKCRL 156
V +V E++R + V+VK RL
Sbjct 133 VHQVVQEVRRTLPGDFSVSVKMRL 156
> CE20317
Length=436
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 79/165 (47%), Gaps = 25/165 (15%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGF------DDV--- 76
+APM+ T R + L YTE +VD ++ T + LG DD+
Sbjct 11 LAPMVRAGRTPLR-LLCLKYGADLCYTEEIVDKKLIEATRVVNEALGTIDYRNGDDIILR 69
Query 77 -----EHPIVCQLGGSNPQNVAEAATWAELAGYD--EVNLNVGCPSCRVVDKGSFGAALM 129
+ + Q+G ++ + +AA A++ G D +++N+GCP + G GAAL+
Sbjct 70 LAPEEKGRCILQIGTNSGE---KAAKIAQIVGDDVAGIDVNMGCPKPFSIHCG-MGAALL 125
Query 130 RSPTKVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETV 174
K+ DI+ +K ++PVT K R+ LD PE T + V+ +
Sbjct 126 TQTEKIVDILTSLKSAAKVPVTCKIRV----LDDPEDTLKLVQEI 166
> Hs22052882
Length=650
Score = 55.5 bits (132), Expect = 6e-08, Method: Composition-based stats.
Identities = 49/150 (32%), Positives = 66/150 (44%), Gaps = 25/150 (16%)
Query 21 RALLAVAPMIAVSNTHFRNFMRLITRRSTVYT--EMVVDSTILHNT---------HQLEN 69
R L +AP+ N FR R+ R T EM V + +L HQ E+
Sbjct 304 RGKLYLAPLTTCGNLPFR---RICKRFGADVTCGEMAVCTNLLQGQMSEWALLKRHQCED 360
Query 70 HLGFDDVEHPIVCQLGGSNPQNVAEAATW-AELAGYDEVNLNVGCPSCRVVDKGSFGAAL 128
G QL G+ P + + A + D V++NVGCP V KG G AL
Sbjct 361 IFG---------VQLEGAFPDTMTKCAELLSRTVEVDFVDINVGCPIDLVYKKGG-GCAL 410
Query 129 MRSPTKVRDIVYEMKRRVQIPVTVKCRLGV 158
M TK + IV M + + +P+TVK R GV
Sbjct 411 MNRSTKFQQIVRGMNQVLDVPLTVKIRTGV 440
> YML080w
Length=423
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 42/159 (26%), Positives = 76/159 (47%), Gaps = 12/159 (7%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRR---STVYTEMVVDSTILHNTHQLENH---LGF 73
R VAPM+ S +R +++RR + YT M+ + E++ L
Sbjct 27 GRPTRIVAPMVDQSELAWR----ILSRRYGATLAYTPMLHAKLFATSKKYREDNWSSLDG 82
Query 74 DDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPT 133
V+ P+V Q ++P+ + AA E D V+LN+GCP + KG +G+ LM
Sbjct 83 SSVDRPLVVQFCANDPEYLLAAAKLVE-DKCDAVDLNLGCPQG-IAKKGHYGSFLMEEWD 140
Query 134 KVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVE 172
+ +++ + + +++PVT K R+ D S + K ++
Sbjct 141 LIHNLINTLHKNLKVPVTAKIRIFDDCEKSLNYAKMVLD 179
> At5g67220
Length=423
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 74/150 (49%), Gaps = 5/150 (3%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENH-LGFDDVEH 78
R VAPM+ S FR + ++ YT M+ S I T + N +
Sbjct 86 GRPKYIVAPMVDNSELPFRLLCQKYGAQA-AYTPML-HSRIFTETEKYRNQEFTTCKEDR 143
Query 79 PIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDI 138
P+ Q ++P + EAA E D V++N+GCP R+ +G++GA LM + V+ +
Sbjct 144 PLFVQFCANDPDTLLEAAKRVE-PYCDYVDINLGCPQ-RIARRGNYGAFLMDNLPLVKSL 201
Query 139 VYEMKRRVQIPVTVKCRLGVDNLDSPEFTK 168
V ++ + + +PV+ K R+ + D+ ++ K
Sbjct 202 VEKLAQNLNVPVSCKIRIFPNLEDTLKYAK 231
> ECU07g1610
Length=341
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 47/157 (29%), Positives = 76/157 (48%), Gaps = 9/157 (5%)
Query 21 RALLAVAPMIAVSNTHFRNFMRLITRRST--VYTEMV-VDSTILHNTHQLENH-LGFDDV 76
R AVAPM+ S +R RL R YTEMV DS + + + ++N +
Sbjct 10 RPYFAVAPMVGNSEEAWR---RLSKRHGANIFYTEMVHCDSFLRGSRNPVKNRWYTTSEG 66
Query 77 EHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVR 136
+ P+V Q+ G++P+ + EAA + D +++N GCP +V KG +GA L +
Sbjct 67 DRPLVVQICGNSPEVMLEAALIIQ-DHCDAIDVNFGCPQ-KVARKGGYGAYLQENWKLTG 124
Query 137 DIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVET 173
+IV + + +PV K R+ + E+ K E
Sbjct 125 EIVKVLSSGLNVPVFCKIRVFGSIEKTVEYAKMIEEA 161
> At3g49640
Length=519
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/174 (24%), Positives = 78/174 (44%), Gaps = 23/174 (13%)
Query 15 ETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGF- 73
+T++ + L +APM+ V FR + Y E ++D ++ +L G
Sbjct 214 KTKMDYQNKLVLAPMVRVGTLSFR-MLAAEYGADITYGEEIIDHKLVKCERRLNVASGTS 272
Query 74 ---------------DDVEHPIVCQLGGSNPQNVAEAATWAELAGYD--EVNLNVGCPSC 116
D+ + +V Q+G S+ +A+ E+ D +++N+GCP
Sbjct 273 EFVEKGTDNVVFSTCDEEKSRVVFQMGTSDAVRALKAS---EIVCNDVATIDINMGCPKA 329
Query 117 RVVDKGSFGAALMRSPTKVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQF 170
+ +G GAAL+ P + DI+ +KR + +PVT K RL D+ E ++
Sbjct 330 FSI-QGGMGAALLSKPELIHDILATLKRNLDVPVTCKIRLLKSPADTVELARRI 382
> Hs5902026
Length=307
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/144 (23%), Positives = 73/144 (50%), Gaps = 6/144 (4%)
Query 19 SSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEH 78
S + + APM+ S FR +R + YT M+V + + + ++ + +
Sbjct 14 SGQLVKVCAPMVRYSKLAFRTLVRKYSC-DLCYTPMIVAADFVKSIKARDSEFTTNQGDC 72
Query 79 PIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDI 138
P++ Q ++ + +++AA + +++N GCP + +G +GA L+ P V+D+
Sbjct 73 PLIVQFAANDARLLSDAARIV-CPYANGIDINCGCPQRWAMAEG-YGACLINKPELVQDM 130
Query 139 VYEMKRRVQIP---VTVKCRLGVD 159
V +++ +V+ P V++K R+ D
Sbjct 131 VKQVRNQVETPGFSVSIKIRINDD 154
> SPBC36B7.04
Length=399
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/155 (26%), Positives = 75/155 (48%), Gaps = 15/155 (9%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTV---YTEMVVDSTILHNTHQLENHLGFDDV---EHP 79
+APM+ S +R ++ RRS Y+ M S + + N + E P
Sbjct 22 LAPMVDQSELPWR----ILARRSGADLCYSPMF-HSRLFGESEDYRNKVFSTRTIPEERP 76
Query 80 IVCQLGGSNPQNVAEAATWAELAGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDI 138
++ Q G++P+ + +AA A A Y D V++N+GCP + KG +G+ L + + I
Sbjct 77 LIIQFCGNDPEIMLKAAKIA--APYCDAVDVNLGCPQG-IAKKGKYGSFLQENWNLIESI 133
Query 139 VYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVET 173
+ ++ + IPVT K R+ D + ++ K ++
Sbjct 134 ITKLHTELSIPVTAKIRIFPDPQKTLDYAKMILKA 168
> SPCC777.15
Length=326
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/131 (28%), Positives = 60/131 (45%), Gaps = 8/131 (6%)
Query 15 ETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFD 74
E R + APM+ S FR +R VYT M++ LH + + +
Sbjct 13 EINKGKRPVHIAAPMVRYSKLPFRQLVRDYNT-DIVYTPMILAKEFLHPKGRYFD-FSTN 70
Query 75 DVEHPIVCQLGGSNPQNVAEAATWAELAG--YDEVNLNVGCPSCRVVDKGSFGAALMRSP 132
D + ++ Q G +P + +AA +L G D + +N GCP + +G G+AL+ P
Sbjct 71 DADASLILQFGVDDPVILEKAA---QLVGPYVDGIGINCGCPQTWAIQEG-IGSALLDEP 126
Query 133 TKVRDIVYEMK 143
KV +V +K
Sbjct 127 EKVHKLVRAVK 137
> CE09942
Length=527
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 71/159 (44%), Gaps = 3/159 (1%)
Query 15 ETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFD 74
ET + R +APM+ S FR F R + T +T M+ +++ N L
Sbjct 66 ETLENQRITKVLAPMVDQSELAFRMFTRKYGAQLT-FTPMIHAHLFVNDGTYRRNSLALV 124
Query 75 DVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
+ P+V Q + A E D V+LN+GCP V +G +G+ L
Sbjct 125 KADRPLVVQFCANKVDTFLAACRLVEDV-CDGVDLNLGCPQM-VAKRGRYGSWLQDEVDL 182
Query 135 VRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVET 173
+ ++V ++ ++P++ K R+ D + E+ K+ V+
Sbjct 183 ICEMVSAVRDYCRLPISCKIRVRDDRQQTVEYAKRLVDA 221
> At4g38890
Length=700
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 70/151 (46%), Gaps = 7/151 (4%)
Query 18 ISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVE 77
I R L +AP+ V N FR +++ T EM + + +L L E
Sbjct 343 IDFRDKLYLAPLTTVGNLPFRRLCKVLGADVTC-GEMAMCTNLLQGQASEWALLRRHSSE 401
Query 78 HPIVCQLGGSNPQNVAEAATWAEL-AGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVR 136
Q+ GS P V+ + D +++N+GCP VV+K S G+AL+ P +++
Sbjct 402 DLFGVQICGSYPDTVSRVVELIDRECTVDFIDINMGCPIDMVVNK-SAGSALLNKPLRMK 460
Query 137 DIVYEMKRRVQIPVTVKCRL----GVDNLDS 163
+IV V+ P+T+K R G + +DS
Sbjct 461 NIVEVSSSIVETPITIKVRTAFFEGKNRIDS 491
> 7292971
Length=460
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/160 (28%), Positives = 68/160 (42%), Gaps = 23/160 (14%)
Query 18 ISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVE 77
+ R L +APM+ V R + L VYTE +VD ++ + + LG D
Sbjct 7 LDYRNKLILAPMVRVGTLPMR-LLALEMGADIVYTEELVDIKLIKSIRRPNPALGTVDFV 65
Query 78 HP----------------IVCQLGGSNPQNVAEAATWAELAGYD--EVNLNVGCPSCRVV 119
P +V Q+G S+ A +L D +++N+GCP +
Sbjct 66 DPSDGTIVFRTCAQETSRLVLQMGTSD---AGRALAVGKLLQRDISGLDINMGCPKEFSI 122
Query 120 DKGSFGAALMRSPTKVRDIVYEMKRRVQIPVTVKCRLGVD 159
KG GAAL+ P K I+ + + IPVT K R+ D
Sbjct 123 -KGGMGAALLADPDKAAHILRTLCSGLDIPVTCKIRILPD 161
> 7296234
Length=358
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 66/137 (48%), Gaps = 9/137 (6%)
Query 26 VAPMIAVSNTHFRNFMRLITRR---STVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVC 82
VAPM+ S +R ++ RR Y+ M + + ++ L + P++
Sbjct 32 VAPMVDQSELAWR----MLCRRYGAELCYSPMYHANLFATDPKYRKDALQTCPEDRPLII 87
Query 83 QLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
Q G++ Q + +AA A+ D V++N+GCP + +G +G+ L + +IV +
Sbjct 88 QFCGNDAQQILDAALLAQ-DHCDAVDINLGCPQA-IAKRGHYGSFLQDEWELLTEIVSTL 145
Query 143 KRRVQIPVTVKCRLGVD 159
++ +PVT K R+ D
Sbjct 146 HAKLAVPVTCKIRIFED 162
> CE01854
Length=290
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/126 (26%), Positives = 59/126 (46%), Gaps = 6/126 (4%)
Query 37 FRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAA 96
FR +R + +T M+ + + + L + + P++ Q +P ++EAA
Sbjct 15 FRQLVR-VYDVDVCFTPMIYAKNFIESEKCRSSELSVCEGDSPLIVQFATDDPFVLSEAA 73
Query 97 TWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEMKRRVQIP---VTVK 153
V+LN GCP V KG FG+AL+ P + D+V + + R+ P V++K
Sbjct 74 EMVYKCSTG-VDLNCGCPKHDVRSKG-FGSALLSKPELLADMVRQTRARIPDPDFSVSLK 131
Query 154 CRLGVD 159
R+ D
Sbjct 132 IRINHD 137
> Hs8923374
Length=493
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 41/150 (27%), Positives = 64/150 (42%), Gaps = 22/150 (14%)
Query 24 LAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP---- 79
L +APM+ V R + L VY E ++D ++ + L D P
Sbjct 14 LILAPMVRVGTLPMR-LLALDYGADIVYCEELIDLKMIQCKRVVNEVLSTVDFVAPDDRV 72
Query 80 -----------IVCQLGGSNPQNVAEAATWAE--LAGYDEVNLNVGCPSCRVVDKGSFGA 126
+V Q+G S+ + A E +AG D +N+GCP + KG GA
Sbjct 73 VFRTCEREQNRVVFQMGTSDAERALAVARLVENDVAGID---VNMGCPK-QYSTKGGMGA 128
Query 127 ALMRSPTKVRDIVYEMKRRVQIPVTVKCRL 156
AL+ P K+ I+ + + + PVT K R+
Sbjct 129 ALLSDPDKIEKILSTLVKGTRRPVTCKIRI 158
> 7298625
Length=604
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 42/145 (28%), Positives = 62/145 (42%), Gaps = 14/145 (9%)
Query 21 RALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPI 80
R L ++P+ + N FR + T EM +L Q E
Sbjct 256 REKLVLSPLTTLGNLPFRRICKEFGADITC-GEMACAQPLLKGMGQEWALTKRHQSEDVF 314
Query 81 VCQLGGSNPQNVAEAAT-WAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIV 139
QL G+NP + +AA E A D ++LN+GCP + +G G+ALMR R +
Sbjct 315 GVQLCGNNPNMLNQAAQVIHETAQVDFIDLNIGCPIDLIYQQGG-GSALMR-----RTNI 368
Query 140 YEMKRRV------QIPVTVKCRLGV 158
E+ R ++P TVK R G+
Sbjct 369 LELTVRSCAALSDRLPFTVKMRTGI 393
> CE19615
Length=554
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 69/151 (45%), Gaps = 11/151 (7%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTVYT--EMVVDSTILHNTHQLENHLGFDDVEHPIVCQ 83
+AP+ V N FR R+ T EM + ++IL T + L E Q
Sbjct 208 LAPLTTVGNLPFR---RICVDYGADITCGEMALATSILSGTASEYSLLKRHPCEKIFGVQ 264
Query 84 LGGSNPQNVAEAATWA-ELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
L G +A+A+ E D +++N+GCP VV++ G AL P K+ +++
Sbjct 265 LAGGFADTMAKASQIVVENFDVDFIDINMGCP-IDVVNQKGGGCALPSRPQKLFEVLAAT 323
Query 143 KRRVQ--IPVTVKCRLGVDN--LDSPEFTKQ 169
K + P+TVK R G+ L +PE+ +
Sbjct 324 KSVLGGCCPLTVKIRTGMKEGVLKAPEYVEH 354
> YLR405w
Length=367
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 61/128 (47%), Gaps = 7/128 (5%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP 79
R + PM+ S FR R VY+ M++ + N H + L ++ + P
Sbjct 37 GRPVTIAGPMVRYSKLPFRQLCREYNV-DIVYSPMILAREYVRNEHARISDLSTNNEDTP 95
Query 80 IVCQLGGSNPQNVAEAATWAEL-AGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRD 137
++ Q+G + NVA+ + E+ A Y D + +N GCP + +G G AL+ + +
Sbjct 96 LIVQVGVN---NVADLLKFVEMVAPYCDGIGINCGCPIKEQIREG-IGCALIYNSDLLCS 151
Query 138 IVYEMKRR 145
+V+ +K +
Sbjct 152 MVHAVKDK 159
> YNR015w
Length=384
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 14/99 (14%)
Query 80 IVCQLGGSNPQNVAEAATWA--ELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRD 137
++ Q+G ++P +AA +++G D +N GCP + G G+AL+R+P +
Sbjct 85 LIFQIGSASPALATQAALKVINDVSGID---INAGCPKHFSIHSG-MGSALLRTPDTLCL 140
Query 138 IVYEMKRRV----QIPVTVKCRLGVDNLDSPEFTKQFVE 172
I+ E+ + V P++VK RL LD+ + T Q V+
Sbjct 141 ILKELVKNVGNPHSKPISVKIRL----LDTKQDTLQLVK 175
> SPAC16.04
Length=617
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 66/153 (43%), Gaps = 16/153 (10%)
Query 18 ISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHL--GFDD 75
I R +AP+ V N FR + T Y+EM + ++ HQ E L G +
Sbjct 235 IDWRDRKILAPLTTVGNPPFRRLCGSLGA-DTFYSEMAMCYPLMQG-HQPEWALVRGLNY 292
Query 76 VEHPIVCQLGGSNPQNVAEAATWA----------ELAGYDEVNLNVGCPSCRVVDKGSFG 125
+ G +A W + G D ++LN GCP V +G+ G
Sbjct 293 EREMMRGGRRGILGVQLATGKLWQATKTAQVIAEQCDGVDFLDLNCGCPIDLVFRQGA-G 351
Query 126 AALMRSPTKV-RDIVYEMKRRVQIPVTVKCRLG 157
++L+ +P ++ R++ QIPVTVK R+G
Sbjct 352 SSLLENPGRLLRNLQGMDAVSGQIPVTVKLRMG 384
> YLR401c
Length=609
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 37/137 (27%), Positives = 60/137 (43%), Gaps = 6/137 (4%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP---IVC 82
V+P+ V N +R MR + T Y+EM + ++ T+ E P +
Sbjct 297 VSPLTTVGNLPYRRLMRKLGADVT-YSEMALAVPLIQGTNSEWALPKAHTSEFPGFGVQV 355
Query 83 QLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
+ A A ++ E+NLN GCP + +GS G+AL+ +P ++ + M
Sbjct 356 ACSKAWQAAKAAEALANSVSEISEINLNSGCPIDLLYRQGS-GSALLDNPARMIRCLNAM 414
Query 143 KR-RVQIPVTVKCRLGV 158
IP+TVK R G
Sbjct 415 NYVSKDIPITVKIRTGT 431
> Hs4503373
Length=1025
Score = 36.6 bits (83), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 59/140 (42%), Gaps = 16/140 (11%)
Query 16 TEISSRALLAV--APMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGF 73
T +S R + PM + F N + LI+ ++ Y + +L+
Sbjct 584 TNVSPRIIRGTTSGPMYGPGQSSFLN-IELISEKTAAY--------WCQSVTELKADFPD 634
Query 74 DDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPT 133
+ V I+C N + E A +E +G D + LN+ CP + + G A + P
Sbjct 635 NIVIASIMCSY---NKNDWTELAKKSEDSGADALELNLSCP--HGMGERGMGLACGQDPE 689
Query 134 KVRDIVYEMKRRVQIPVTVK 153
VR+I +++ VQIP K
Sbjct 690 LVRNICRWVRQAVQIPFFAK 709
> CE02496
Length=1084
Score = 32.7 bits (73), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 2/66 (3%)
Query 88 NPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEMKRRVQ 147
N + E AT +E AG D + LN+ CP + +KG G A +SP V++I ++ V+
Sbjct 685 NKADWIELATKSEEAGADILELNLSCPH-GMGEKG-MGLACGQSPEIVKEICRWVRACVK 742
Query 148 IPVTVK 153
IP K
Sbjct 743 IPFFPK 748
> 7303851
Length=1127
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 21/77 (27%), Positives = 35/77 (45%), Gaps = 6/77 (7%)
Query 102 AGYDEVNLNVGCPSCRV------VDKGSFGAALMRSPTKVRDIVYEMKRRVQIPVTVKCR 155
A +V PSCR V+ S L ++P+ D++ E K+ V+ VK
Sbjct 261 ASASDVKCACDAPSCRFMESSKQVEPTSPTPTLPKAPSSELDVIREYKQAVEGVQVVKNH 320
Query 156 LGVDNLDSPEFTKQFVE 172
LG D L++ E +++
Sbjct 321 LGTDTLNNIEILPNYLD 337
> CE15837
Length=337
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 20/27 (74%), Gaps = 2/27 (7%)
Query 53 EMVVDSTILHNTHQLENH--LGFDDVE 77
++V + T+LH T+ LEN+ LGF DV+
Sbjct 171 KLVAELTVLHPTYGLENYAILGFSDVK 197
> YOR056c
Length=459
Score = 28.9 bits (63), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 42/84 (50%), Gaps = 11/84 (13%)
Query 21 RAL-LAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP 79
RAL L P+I S TH++N+ + TV+ E + D+ N ++ LG + HP
Sbjct 10 RALILDATPLITQSYTHYQNYAQSFYTTPTVFQE-IKDAQARKNL-EIWQSLGTLKLVHP 67
Query 80 IVCQLGGSNPQNVAEAATWAELAG 103
+ ++A+ +T+A+L G
Sbjct 68 --------SENSIAKVSTFAKLTG 83
> YDR034c-d
Length=1770
Score = 28.5 bits (62), Expect = 7.5, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YOR192c-b
Length=1770
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YLR410w-b
Length=1770
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YOR343c-b
Length=1770
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YDR261w-b
Length=1770
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YDR210w-b
Length=1770
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YCL019w
Length=1770
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YBL101w-b
Length=1770
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YGR161w-b
Length=1770
Score = 28.5 bits (62), Expect = 8.1, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
> YFL002w-a
Length=1770
Score = 28.5 bits (62), Expect = 8.1, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 10/91 (10%)
Query 59 TILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYD---------EVNL 109
T+LH + L NHL F VE + + +P+N A A LAG D V +
Sbjct 780 TLLHCS-GLPNHLWFSAVEFSTIIRNSLVSPKNDKSARQHAGLAGLDITTILPFGQPVIV 838
Query 110 NVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
N P ++ +G G AL S I+Y
Sbjct 839 NNHNPDSKIHPRGIPGYALHPSRNSYGYIIY 869
Lambda K H
0.320 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2671071884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40