bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1936_orf1
Length=256
Score E
Sequences producing significant alignments: (Bits) Value
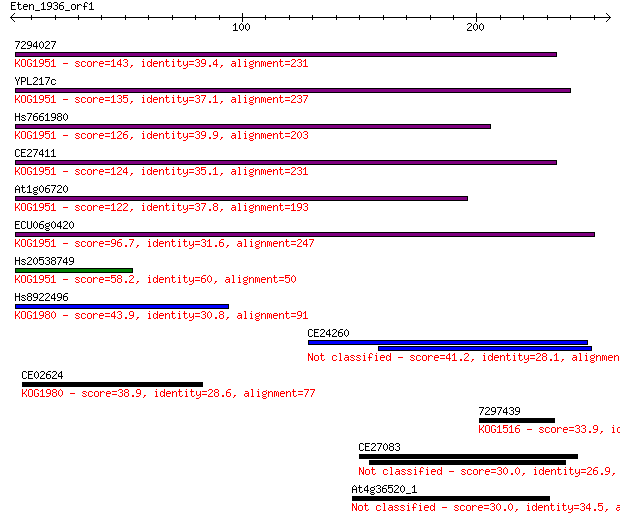
7294027 143 3e-34
YPL217c 135 7e-32
Hs7661980 126 6e-29
CE27411 124 2e-28
At1g06720 122 8e-28
ECU06g0420 96.7 5e-20
Hs20538749 58.2 2e-08
Hs8922496 43.9 4e-04
CE24260 41.2 0.002
CE02624 38.9 0.010
7297439 33.9 0.33
CE27083 30.0 4.7
At4g36520_1 30.0 5.3
> 7294027
Length=1159
Score = 143 bits (361), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 91/240 (37%), Positives = 146/240 (60%), Gaps = 20/240 (8%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGT-DGTFRAS 61
KLKLVG I +KTAFVK MFNS LEV + GAKI+TVSGIRGQIKKA T +G++RA+
Sbjct 916 KLKLVGHPFKIYKKTAFVKDMFNSSLEVAKFEGAKIKTVSGIRGQIKKAHHTPEGSYRAT 975
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPV--LDLP-----GWRGLRLLSEIKKEKKI-ISK 113
FEDK+L SD++ C+TW V FY PV L LP W+G++ L ++K+E+ + +
Sbjct 976 FEDKILLSDIVFCRTWFRVEVPRFYAPVSSLLLPLDQKSQWQGMKTLGQLKRERAVQNAA 1035
Query 114 QKETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKA 173
Q ++++ + RK+ F P+ IPK L LP + KL G + + +
Sbjct 1036 QPDSMYTTI---VRKEKIFRPLTIPKALQRALPYKDKPKL------GPENPKAALERVAV 1086
Query 174 FVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQKRR 233
SP E++ + +++ +ET +DKR++++ + K R+K + K ++ Q++++++ R+
Sbjct 1087 VNSPYEQKVSKMMKMIET--NFKDKRQRERMEMKKRIKNYREKKREKMASQERRQKELRK 1144
> YPL217c
Length=1183
Score = 135 bits (340), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 88/247 (35%), Positives = 149/247 (60%), Gaps = 21/247 (8%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIG-TDGTFRAS 61
KLKLVG I + TAF+K MF+S +EV R GA+I+TVSGIRG+IK+A+ +G +RA+
Sbjct 931 KLKLVGFPYKIFKNTAFIKDMFSSAMEVARFEGAQIKTVSGIRGEIKRALSKPEGHYRAA 990
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPVLDL-----PGWRGLRLLSEIKKEKKI-ISKQK 115
FEDK+L SD+++ ++W V FYNPV L W+GLRL +I+ +
Sbjct 991 FEDKILMSDIVILRSWYPVRVKKFYNPVTSLLLKEKTEWKGLRLTGQIRAAMNLETPSNP 1050
Query 116 ETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAFV 175
++ +H K R + F +++PK + +LP S++ K Q++K K+A V
Sbjct 1051 DSAYH---KIERVERHFNGLKVPKAVQKELPFKSQIHQM------KPQKKKTYMAKRAVV 1101
Query 176 -SPNERRAAVLLQQLETLKQHRDKRRQQQQ--QQKARLKQQQTDKLQQQQQQQQKERQKR 232
+E++A +Q++ T+ + +D +R++Q+ Q+K RLK+ K+++++ Q+ KE++K
Sbjct 1102 LGGDEKKARSFIQKVLTISKAKDSKRKEQKASQRKERLKKLA--KMEEEKSQRDKEKKKE 1159
Query 233 RHSKQGK 239
++ GK
Sbjct 1160 YFAQNGK 1166
> Hs7661980
Length=1282
Score = 126 bits (316), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 81/212 (38%), Positives = 117/212 (55%), Gaps = 16/212 (7%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI-GTDGTFRAS 61
KLKL G I + T+F+KGMFNS LEV + GA I+TVSGIRGQIKKA+ +G FRAS
Sbjct 1026 KLKLTGFPYKIFKNTSFIKGMFNSALEVAKFEGAVIRTVSGIRGQIKKALRAPEGAFRAS 1085
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPVLDL-------PGWRGLRLLSEIKKEKKI-ISK 113
FEDK+L SD++ +TW V AFYNPV L W G+R +++ + +
Sbjct 1086 FEDKLLMSDIVFMRTWYPVSIPAFYNPVTSLLKPVGEKDTWSGMRTTGQLRLAHGVRLKA 1145
Query 114 QKETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKA 173
K++L+ R++ F + IPK L LP ++ K Q + GK + + R
Sbjct 1146 NKDSLYKPI---LRQKKHFNSLHIPKALQKALPFKNKPKTQA--KAGKVPKDRRR--PAV 1198
Query 174 FVSPNERRAAVLLQQLETLKQHRDKRRQQQQQ 205
P+ER+ LL L T+ + K+ ++Q+
Sbjct 1199 IREPHERKILALLDALSTVHSQKMKKAKEQRH 1230
> CE27411
Length=920
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 81/245 (33%), Positives = 133/245 (54%), Gaps = 29/245 (11%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI-GTDGTFRAS 61
KLKL+G + I +KTAFVKGMFNS LEV + GA I+TV+GIRGQIKKAI +G FRA+
Sbjct 665 KLKLIGHPEKIFKKTAFVKGMFNSALEVAKFEGATIRTVAGIRGQIKKAIKAPEGAFRAT 724
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPVLD-----LPGWRGLRLLSEIKKEKKIISKQKE 116
FEDK+L D++ ++WV V FY P+ D W G+R + +++ E + + Q +
Sbjct 725 FEDKILMRDIVFLRSWVTVPIPRFYTPISDHLQASAAAWIGMRTVGKMRSELGMGTPQNK 784
Query 117 TLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAFVS 176
+ RK+ PI +P +L LP + Q +++KE+D A
Sbjct 785 D--SDYKPIVRKEFESAPIHLPPKLQKSLPFKMKPTYQ-------AREEKEKDSLVA--- 832
Query 177 PNERRAAVLLQQ--------LETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKE 228
R AV+L+ ++ L+ D ++Q+ K K+++ +++ + + +++K
Sbjct 833 ---RHTAVVLEPEEAKRERFMDMLRTLNDVDLKKQENIKEGYKKRKAEEMAESEAKREKS 889
Query 229 RQKRR 233
+ R+
Sbjct 890 IKSRK 894
> At1g06720
Length=1147
Score = 122 bits (306), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 73/206 (35%), Positives = 113/206 (54%), Gaps = 21/206 (10%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI-------GTD 55
K+KLVG IK+KTAF+K MF S LE+ R G+ ++TVSGIRGQ+KKA +
Sbjct 896 KIKLVGTPCKIKKKTAFIKDMFTSDLEIARFEGSSVRTVSGIRGQVKKAGKNMLDNKAEE 955
Query 56 GTFRASFEDKVLKSDLILCKTWVLVCPHAFYNPVLDL-----PGWRGLRLLSEIKKEKKI 110
G R +FED++ SD++ + W V FYNP+ W G++ E+++E I
Sbjct 956 GIARCTFEDQIHMSDMVFLRAWTTVEVPQFYNPLTTALQPRDKTWNGMKTFGELRRELNI 1015
Query 111 -ISKQKETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERD 169
I K++L+ + RKQ F P++IPK+L LP S+ K + K+++ D
Sbjct 1016 PIPVNKDSLYKAI---ERKQKKFNPLQIPKRLEKDLPFMSKPK-----NIPKRKRPSLED 1067
Query 170 MKKAFVSPNERRAAVLLQQLETLKQH 195
+ + P ER+ ++QQ + L+ H
Sbjct 1068 KRAVIMEPKERKEHTIIQQFQLLQHH 1093
> ECU06g0420
Length=777
Score = 96.7 bits (239), Expect = 5e-20, Method: Composition-based stats.
Identities = 78/251 (31%), Positives = 129/251 (51%), Gaps = 27/251 (10%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
KLKLVG K I + T FV+ MF S LEV + GA ++ VSG+RGQ+K G +G +RA F
Sbjct 547 KLKLVGYPKQIVQNTVFVRDMFTSDLEVLKFEGASLKAVSGLRGQVKGPHGKNGEYRAVF 606
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLPG-WRGLRLLSEIKKEKKII---SKQKETL 118
E K+L SD+I + +V V H + PV +L G WRGLR L EI++ + + Q ++
Sbjct 607 EGKMLMSDIITLRCFVPVEVHRIFIPVDNLLGKWRGLRRLHEIRESLGLTHSYAPQNDSS 666
Query 119 FHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAFVSPN 178
G + +S +P+++ +KLPL R RR+ P
Sbjct 667 SEEMGYGAEEDYS-----LPREIESKLPLDKRSIAVVSRRI-------------ELPVPP 708
Query 179 ERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQKRRHSKQG 238
E R ++ E + +R ++ Q++K R++ Q K ++ +++++ Q+ R +
Sbjct 709 ECR-----EKHEIKDRIVKERIRKDQEEKERMESLQRAKEEEIGKKEKEREQRIRKTIHD 763
Query 239 KIEASMRKRLR 249
+ +KRL+
Sbjct 764 NYKEMAKKRLK 774
> Hs20538749
Length=192
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/50 (60%), Positives = 37/50 (74%), Gaps = 0/50 (0%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI 52
KLKL G I + T+F+KGMFNS LEV + A I+TVSGIRGQIK+A+
Sbjct 143 KLKLTGFPYKIFKNTSFIKGMFNSALEVAKFEDAVIRTVSGIRGQIKRAL 192
> Hs8922496
Length=656
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 45/91 (49%), Gaps = 0/91 (0%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
++ L G I K A V+ MF + +V +++T G RG IK+ +GT G + SF
Sbjct 548 RVVLSGHPFKIFTKMAVVRYMFFNREDVLWFKPVELRTKWGRRGHIKEPLGTHGHMKCSF 607
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLP 93
+ K+ D +L + V P Y+P + P
Sbjct 608 DGKLKSQDTVLMNLYKRVFPKWTYDPYVPEP 638
> CE24260
Length=923
Score = 41.2 bits (95), Expect = 0.002, Method: Composition-based stats.
Identities = 33/135 (24%), Positives = 68/135 (50%), Gaps = 20/135 (14%)
Query 128 KQHSFFPIRIPKQLAAKLPLHSRLKLQQ-------------GRRVGKQQQQKERDMKKAF 174
K H+ FP +IP ++ + L + QQ + +Q Q++E+ + + F
Sbjct 181 KNHNRFPKQIPVAISTREELQQQRHKQQLEEYEKRKEILRREEEIKQQNQEREKQINEDF 240
Query 175 VSPNERRAAVLLQQLETLKQHRDKRRQQQQQ--QKARLKQQQTDKLQQQQQQQQKERQK- 231
E+R L++ E +KQ +R +Q +K +L++ Q +KL ++ +Q + +R+K
Sbjct 241 ----EKRNQEKLKREEEIKQQNQEREKQINDDFEKRKLEKLQREKLFEKNKQAKLQREKA 296
Query 232 RRHSKQGKIEASMRK 246
+ S++ K+E R+
Sbjct 297 HQESERRKLEKLKRE 311
Score = 29.3 bits (64), Expect = 9.1, Method: Composition-based stats.
Identities = 27/105 (25%), Positives = 51/105 (48%), Gaps = 14/105 (13%)
Query 158 RVGKQQQQKERDMKKAFVSPNERRAAVLLQQLETLKQHRDKRRQQQ---QQQKARLKQQQ 214
RV K + + + A + E + QQLE ++ ++ R+++ QQ + R KQ
Sbjct 178 RVIKNHNRFPKQIPVAISTREELQQQRHKQQLEEYEKRKEILRREEEIKQQNQEREKQIN 237
Query 215 TD-------KLQQQ----QQQQQKERQKRRHSKQGKIEASMRKRL 248
D KL+++ QQ Q++E+Q ++ K+E R++L
Sbjct 238 EDFEKRNQEKLKREEEIKQQNQEREKQINDDFEKRKLEKLQREKL 282
> CE02624
Length=785
Score = 38.9 bits (89), Expect = 0.010, Method: Composition-based stats.
Identities = 22/77 (28%), Positives = 38/77 (49%), Gaps = 0/77 (0%)
Query 6 LVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASFEDK 65
L G I R+ V+ MF + ++ ++ T SG RG IK+A+GT G + F+ +
Sbjct 674 LAGHPYKINRRAVVVRYMFFNREDIEWFKPVELYTPSGRRGHIKEAVGTHGNMKCRFDQQ 733
Query 66 VLKSDLILCKTWVLVCP 82
+ D ++ + V P
Sbjct 734 LNAQDSVMLNLYKRVFP 750
> 7297439
Length=1026
Score = 33.9 bits (76), Expect = 0.33, Method: Composition-based stats.
Identities = 11/32 (34%), Positives = 27/32 (84%), Gaps = 0/32 (0%)
Query 201 QQQQQQKARLKQQQTDKLQQQQQQQQKERQKR 232
++QQ ++ R +++Q ++L+QQ++Q+++ RQ+R
Sbjct 681 REQQDERIRQQREQEERLRQQREQEERLRQQR 712
> CE27083
Length=1480
Score = 30.0 bits (66), Expect = 4.7, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 53/94 (56%), Gaps = 12/94 (12%)
Query 150 RLKLQQGRRVGKQQQQKERDMKKAFVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKAR 209
R++ Q RR ++Q +++R+ N RR +L Q E + R R QQ ++A
Sbjct 270 RIREDQARRQQEEQDRRDRE-------DNARR---ILAQREHQEMER-LREQQNLSERAL 318
Query 210 LKQQQTDKLQQQQQQQ-QKERQKRRHSKQGKIEA 242
++++ DK + QQ++ +++R+K+R + ++E+
Sbjct 319 AERERADKERLQQERLLRQQREKKRREEWDRLES 352
Score = 29.6 bits (65), Expect = 5.6, Method: Composition-based stats.
Identities = 20/84 (23%), Positives = 44/84 (52%), Gaps = 5/84 (5%)
Query 154 QQGRRVGKQQQQKERDMKKAFVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQ 213
Q+ R+ +QQ ER + + + ER LQQ L+Q R+K+R+++ + ++
Sbjct 302 QEMERLREQQNLSERALAERERADKER-----LQQERLLRQQREKKRREEWDRLESIRLA 356
Query 214 QTDKLQQQQQQQQKERQKRRHSKQ 237
+ + +++ +KER R +++
Sbjct 357 EEEAELARRRALEKERIDREKAEE 380
> At4g36520_1
Length=1289
Score = 30.0 bits (66), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 53/89 (59%), Gaps = 6/89 (6%)
Query 147 LHSRLKLQQGRRVGKQQQQKERDMKKAFVSPNERR----AAVLLQQLE-TLKQHRDKRRQ 201
+ +R K +Q R++ K+QQ+ E +K+AF E R A L Q+ E +K+ R+K
Sbjct 691 VEAREKAEQERKM-KEQQELELQLKEAFEKEEENRRMREAFALEQEKERRIKEAREKEEN 749
Query 202 QQQQQKARLKQQQTDKLQQQQQQQQKERQ 230
+++ ++AR K + +L+ +Q++KERQ
Sbjct 750 ERRIKEAREKAELEQRLKATLEQEEKERQ 778
Lambda K H
0.319 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5329644900
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40