bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1964_orf4
Length=347
Score E
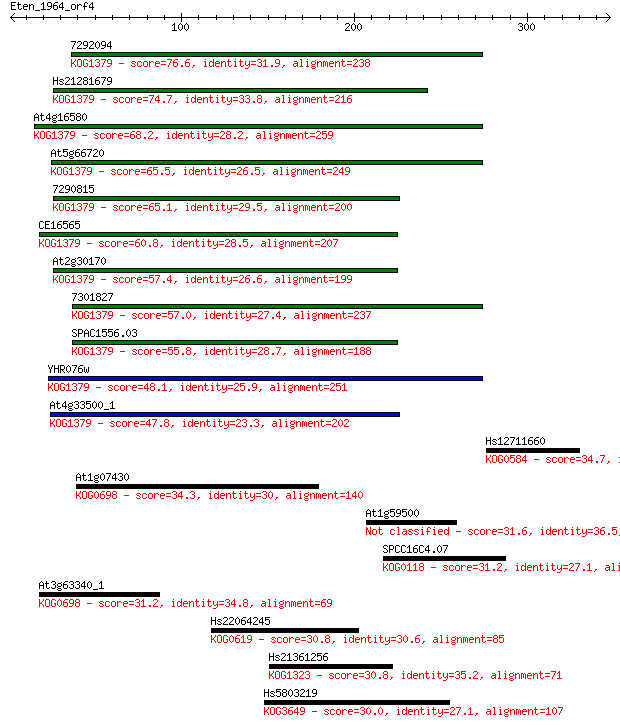
Sequences producing significant alignments: (Bits) Value
7292094 76.6 6e-14
Hs21281679 74.7 3e-13
At4g16580 68.2 2e-11
At5g66720 65.5 2e-10
7290815 65.1 2e-10
CE16565 60.8 5e-09
At2g30170 57.4 4e-08
7301827 57.0 6e-08
SPAC1556.03 55.8 1e-07
YHR076w 48.1 3e-05
At4g33500_1 47.8 4e-05
Hs12711660 34.7 0.32
At1g07430 34.3 0.38
At1g59500 31.6 2.9
SPCC16C4.07 31.2 3.1
At3g63340_1 31.2 3.3
Hs22064245 30.8 4.2
Hs21361256 30.8 4.5
Hs5803219 30.0 7.0
> 7292094
Length=321
Score = 76.6 bits (187), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 76/238 (31%), Positives = 102/238 (42%), Gaps = 71/238 (29%)
Query 36 FGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSLVKKVKEMQHGFNVPF 95
GSSTA V +L+ E+ AN+GDS +V+R +V V K +E QH FN PF
Sbjct 153 LGSSTACVLILNRETSTVHTANIGDSGFVVVREGQV---------VHKSEEQQHYFNTPF 203
Query 96 QFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAIQSTSVPVQDGDLIIL 155
Q + LP P +L D P + + S PV+DGD+I++
Sbjct 204 QLS-LPPPGHGPNVLS-----------------------DSPESADTMSFPVRDGDVILI 239
Query 156 GTDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPEDVARALALAAYWRSLDRDA 215
TDG+FDNV E L L L+ E + L + A +LAL A SL+ +
Sbjct 240 ATDGVFDNV--PEDLMLQVLS----EVEGERDPVKL----QMTANSLALMARTLSLNSEF 289
Query 216 QTPFSKEARRCCNKQSLPVQTPDGSTGAHQASPAHAMFLAGLLSGGKEDDITVAAAWV 273
+PF+ ARR N Q+ GGK DDITV A V
Sbjct 290 LSPFALSARR-NNIQA---------------------------RGGKPDDITVVLATV 319
> Hs21281679
Length=304
Score = 74.7 bits (182), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 73/222 (32%), Positives = 98/222 (44%), Gaps = 53/222 (23%)
Query 26 LTRAY-----HKARNFGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSL 80
LT +Y +K GSSTA + VLD S ANLGDS LV+R V
Sbjct 119 LTTSYCELLQNKVPLLGSSTACIVVLDRTSHRLHTANLGDSGFLVVRGGEV--------- 169
Query 81 VKKVKEMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAI 140
V + E QH FN PFQ + P PE +G + D P A
Sbjct 170 VHRSDEQQHYFNTPFQLSIAP-PE-----------------------AEGVVLSDSPDAA 205
Query 141 QSTSVPVQDGDLIILGTDGLFDNVFDFEAL-ALTGLAVSPHEAQTLLGDASLATPPEDVA 199
STS VQ GD+I+ TDGLFDN+ D+ L L L S +E+ + A
Sbjct 206 DSTSFDVQLGDIILTATDGLFDNMPDYMILQELKKLKNSNYES------------IQQTA 253
Query 200 RALALAAYWRSLDRDAQTPFSKEARRCCNKQSLPVQTPDGST 241
R++A A+ + D + +PF++ A C N ++ PD T
Sbjct 254 RSIAEQAHELAYDPNYMSPFAQFA--CDNGLNVRGGKPDDIT 293
> At4g16580
Length=335
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 73/260 (28%), Positives = 105/260 (40%), Gaps = 88/260 (33%)
Query 15 TCNPADVAKEALTRAYHKARNFGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGS 74
+ +PA V L +A+ ++ GSSTA + L + G+ + NLGDS +V+R
Sbjct 154 SIDPARV----LEKAHTCTKSQGSSTACIIALTNQ-GLHAI-NLGDSGFMVVREGHT--- 204
Query 75 LGQLSLVKKVKEMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIG 134
V + QH FN +Q + GR G
Sbjct 205 ------VFRSPVQQHDFNFTYQL-------------------------------ESGRNG 227
Query 135 DEPSAIQSTSVPVQDGDLIILGTDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATP 194
D PS+ Q +V V GD+II GTDGLFDN+++ E A+ AV +
Sbjct 228 DLPSSGQVFTVAVAPGDVIIAGTDGLFDNLYNNEITAIVVHAVRAN------------ID 275
Query 195 PEDVARALALAAYWRSLDRDAQTPFSKEARRCCNKQSLPVQTPDGSTGAHQASPAHAMFL 254
P+ A+ +A A R+ D++ QTPFS A+
Sbjct 276 PQVTAQKIAALARQRAQDKNRQTPFSTAAQD----------------------------- 306
Query 255 AGL-LSGGKEDDITVAAAWV 273
AG GGK DDITV ++V
Sbjct 307 AGFRYYGGKLDDITVVVSYV 326
> At5g66720
Length=414
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 66/249 (26%), Positives = 104/249 (41%), Gaps = 82/249 (32%)
Query 25 ALTRAYHKARNFGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSLVKKV 84
L +A+ + + GSSTA + VL + G+ + NLGDS V+R + + V +
Sbjct 245 VLEKAHSQTKAKGSSTACIIVLK-DKGLHAI-NLGDSGFTVVR---------EGTTVFQS 293
Query 85 KEMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAIQSTS 144
QHGFN +Q + G D PS+ Q +
Sbjct 294 PVQQHGFNFTYQL-------------------------------ESGNSADVPSSGQVFT 322
Query 145 VPVQDGDLIILGTDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPEDVARALAL 204
+ VQ GD+I+ GTDG++DN+++ E +TG+ VS A P+ A+ +A
Sbjct 323 IDVQSGDVIVAGTDGVYDNLYNEE---ITGVVVSSVRA---------GLDPKGTAQKIAE 370
Query 205 AAYWRSLDRDAQTPFSKEARRCCNKQSLPVQTPDGSTGAHQASPAHAMFLAGLLSGGKED 264
A R++D+ Q+PF +T A +A + GGK D
Sbjct 371 LARQRAVDKKRQSPF--------------------ATAAQEAGYRYY--------GGKLD 402
Query 265 DITVAAAWV 273
DIT ++V
Sbjct 403 DITAVVSYV 411
> 7290815
Length=374
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 59/208 (28%), Positives = 82/208 (39%), Gaps = 54/208 (25%)
Query 26 LTRAY-----HKARNFGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSL 80
L RAY K GS TA + L + AN+GDS LV+R +V
Sbjct 192 LERAYFDLLDQKCPIVGSCTACILALKRDDSTLYAANIGDSGFLVVRSGKV--------- 242
Query 81 VKKVKEMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAI 140
V + +E QH FN P+Q A P ++D + D P +
Sbjct 243 VCRSQEQQHQFNTPYQLASPPPGYDFDA------------------------VSDGPESA 278
Query 141 QSTSVPVQDGDLIILGTDGLFDNV---FDFEALALTGLAVSPHEAQTLLGDASLATPPED 197
+ P+Q GD+I+L TDG++DNV F E L +P Q
Sbjct 279 DTIQFPMQLGDVILLATDGVYDNVPESFLVEVLTEMSGISNPVRLQM------------- 325
Query 198 VARALALAAYWRSLDRDAQTPFSKEARR 225
A +AL A S +PFS+ AR+
Sbjct 326 AANTVALMARTLSFSPKHDSPFSQNARK 353
> CE16565
Length=330
Score = 60.8 bits (146), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 59/208 (28%), Positives = 83/208 (39%), Gaps = 50/208 (24%)
Query 18 PADVAKEALTRAYHKARNFGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQ 77
P + A + R GSSTA V V+ E ANLGDS +V+R ++
Sbjct 150 PESLLDYAFRASAEAPRPVGSSTACVLVVHQEK--LYSANLGDSGFMVVRNGKI------ 201
Query 78 LSLVKKVKEMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEP 137
V K +E H FN PFQ P +G G IGD+
Sbjct 202 ---VSKSREQVHYFNAPFQLTLPP-------------------------EGYQGFIGDKA 233
Query 138 SAIQSTSVPVQDGDLIILGTDGLFDNVFDFEAL-ALTGLAVSPHEAQTLLGDASLATPPE 196
+ V+ GD+I+L TDG++DN+ + + L L L Q
Sbjct 234 DMADKDEMAVKKGDIILLATDGVWDNLSEQQVLDQLKALDAGKSNVQ------------- 280
Query 197 DVARALALAAYWRSLDRDAQTPFSKEAR 224
+V ALAL A + D +PF+ +AR
Sbjct 281 EVCNALALTARRLAFDSKHNSPFAMKAR 308
> At2g30170
Length=283
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 53/199 (26%), Positives = 85/199 (42%), Gaps = 44/199 (22%)
Query 26 LTRAYHKARNFGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSLVKKVK 85
+ +A+ + GS+T + + E GI + N+GD +LR GQ+ +
Sbjct 122 IDKAHTATTSRGSATISILAMLEEVGILKIGNVGDCGLKLLRE-------GQIIFATAPQ 174
Query 86 EMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAIQSTSV 145
E H F+ P+Q + +G D A Q + V
Sbjct 175 E--HYFDCPYQLS---------------------------SEGSAQTYLD---ASQFSIV 202
Query 146 PVQDGDLIILGTDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPEDVARALALA 205
VQ GD+I++G+DGLFDNVFD E +++ E+ + + T AR LA
Sbjct 203 EVQKGDVIVMGSDGLFDNVFDHEIVSIVTKHTDVAESLSYMFFGITIT-----ARLLAEV 257
Query 206 AYWRSLDRDAQTPFSKEAR 224
A S D + ++P++ EAR
Sbjct 258 ASSHSRDTEFESPYALEAR 276
> 7301827
Length=314
Score = 57.0 bits (136), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 65/237 (27%), Positives = 98/237 (41%), Gaps = 71/237 (29%)
Query 37 GSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSLVKKVKEMQHGFNVPFQ 96
GSSTA + + + ANLGDS LV+R RV + + E H FN P+Q
Sbjct 144 GSSTACLATMHRKDCTLYTANLGDSGFLVVRNGRV---------LHRSVEQTHDFNTPYQ 194
Query 97 FAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAIQSTSVPVQDGDLIILG 156
+P PE+ K+ Y D+P ST + GDL++L
Sbjct 195 LT-VP-PED---------------RKESY-------YCDKPEMAVSTRHSLLPGDLVLLA 230
Query 157 TDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPEDVARALALAAYWRSLDRDAQ 216
TDGLFDN+ + L++ E L+G + + + AR L++ A + Q
Sbjct 231 TDGLFDNMPESMLLSILNGLKERGEHDLLVGASRVV----EKARELSMNASF-------Q 279
Query 217 TPFSKEARRCCNKQSLPVQTPDGSTGAHQASPAHAMFLAGLLSGGKEDDITVAAAWV 273
+PF+ +AR+ H + +G GGK DDIT+ + V
Sbjct 280 SPFAIKARQ------------------------HNVSYSG---GGKPDDITLILSSV 309
> SPAC1556.03
Length=288
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 54/190 (28%), Positives = 82/190 (43%), Gaps = 45/190 (23%)
Query 37 GSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSLVKKVKEMQHGFNVPFQ 96
GSSTA + + + +G NLGDS L+LR G + + +Q FN+P+Q
Sbjct 126 GSSTACLTLFNCGNGKLHSLNLGDSGFLILRN-------GAIHYASPAQVLQ--FNMPYQ 176
Query 97 FAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAIQSTSVPVQDGDLIILG 156
A P + Y + IG P Q+T ++D DL+IL
Sbjct 177 LAIYP---------------------RNYRSAE--NIG--PKMGQATVHDLKDNDLVILA 211
Query 157 TDGLFDNVFDFEALALTGLA--VSPHEAQTLLGDASLATPPEDVARALALAAYWRSLDRD 214
TDG+FDN+ + L + G+ S Q L +D+A + A SLD
Sbjct 212 TDGIFDNIEEKSILDIAGVVDFSSLSNVQKCL---------DDLAMRICRQAVLNSLDTK 262
Query 215 AQTPFSKEAR 224
++PF+K A+
Sbjct 263 WESPFAKTAK 272
> YHR076w
Length=374
Score = 48.1 bits (113), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 65/258 (25%), Positives = 96/258 (37%), Gaps = 74/258 (28%)
Query 23 KEALTRAYHKARN-----FGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQ 77
K+ + AY K R+ G +TAIV +G +ANLGDS V R +
Sbjct 183 KKIIGAAYAKIRDEKVVKVGGTTAIVAHFPS-NGKLEVANLGDSWCGVFRDSK------- 234
Query 78 LSLVKKVKEMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEP 137
LV + K GFN P+Q + +P +E+L+ E ++ I + P
Sbjct 235 --LVFQTKFQTVGFNAPYQLSIIP-----EEMLK------------EAERRGSKYILNTP 275
Query 138 SAIQSTSVPVQDGDLIILGTDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPED 197
S ++ D+IIL TDG+ DN ++ + + L D + T E
Sbjct 276 RDADEYSFQLKKKDIIILATDGVTDN-------------IATDDIELFLKDNAARTNDEL 322
Query 198 VARALALAAYWRSLDRDAQTP--FSKEARRCCNKQSLPVQTPDGSTGAHQASPAHAMFLA 255
+ SL +D P F++E + K
Sbjct 323 QLLSQKFVDNVVSLSKDPNYPSVFAQEISKLTGKN------------------------- 357
Query 256 GLLSGGKEDDITVAAAWV 273
SGGKEDDITV V
Sbjct 358 --YSGGKEDDITVVVVRV 373
> At4g33500_1
Length=702
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 47/202 (23%), Positives = 82/202 (40%), Gaps = 56/202 (27%)
Query 24 EALTRAYHKARNFGSSTAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSLVKK 83
+ L R+ ++ ++ GSSTA++ LD +AN+GDS +V+R G++ Q S
Sbjct 536 QVLHRSVNETKSSGSSTALIAHLDNNE--LHIANIGDSGFMVIRD----GTVLQNS---- 585
Query 84 VKEMQHGFNVPFQFAHLPAPEEWDELLQRGLTRLVSIAKQEYDQGDGGRIGDEPSAIQST 143
M H F P QG D +
Sbjct 586 -SPMFHHFCFPLHIT----------------------------QG-----CDVLKLAEVY 611
Query 144 SVPVQDGDLIILGTDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPEDVARALA 203
V +++GD++I TDGLFDN+++ E +++ + G + P+ +A +A
Sbjct 612 HVNLEEGDVVIAATDGLFDNLYEKEIVSI------------VCGSLKQSLEPQKIAELVA 659
Query 204 LAAYWRSLDRDAQTPFSKEARR 225
A + +TPF+ A+
Sbjct 660 AKAQEVGRSKTERTPFADAAKE 681
> Hs12711660
Length=2382
Score = 34.7 bits (78), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 276 KDRCVYTEN-DGKQSSPPYTRREAQAGQFRRPQQQQCQQASQGEDLKTKKTGLSN 329
KDR V + G + PP R + G + PQ+++ QQ E+L+TK G+SN
Sbjct 162 KDRPVSQPSLVGSKEEPPPARSGSGGGSAKEPQEERSQQQDDIEELETKAVGMSN 216
> At1g07430
Length=442
Score = 34.3 bits (77), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 42/158 (26%), Positives = 63/158 (39%), Gaps = 50/158 (31%)
Query 39 STAIVGVLDGESGIFGLANLGDSSSLVLRRPRVRGSLGQLSLVKKVKEMQHGFNVPFQFA 98
STA+V V+ E I +AN GDS +++ R +G VP
Sbjct 234 STAVVSVITPEKII--VANCGDSRAVLCR---------------------NGKAVPLSTD 270
Query 99 HLP-APEEWDELLQRGLTRLVSIAKQEYDQGDGGR----------IGD---EPSAIQSTS 144
H P P+E D + + G R++ DG R IGD +P
Sbjct 271 HKPDRPDELDRIQEAG-GRVI--------YWDGARVLGVLAMSRAIGDNYLKPYVTSEPE 321
Query 145 VPVQD----GDLIILGTDGLFDNVFDFEALALTGLAVS 178
V V D + +IL TDGL+D V + A + + ++
Sbjct 322 VTVTDRTEEDEFLILATDGLWDVVTNEAACTMVRMCLN 359
> At1g59500
Length=597
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 207 YWRSLDRDAQTPFSKEARRCCN-KQSLPVQTPDGSTGAHQASPAHAMFLAGLL 258
Y+R+ D D+ KEA CC+ QS+ Q G H+ + A+F +GLL
Sbjct 182 YFRTSDSDSVYTSPKEAILCCDSSQSMYTQMLCGLLMRHEVNRLGAVFPSGLL 234
> SPCC16C4.07
Length=561
Score = 31.2 bits (69), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 32/71 (45%), Gaps = 1/71 (1%)
Query 217 TPFSKEARRCCNKQSLPVQTPDGSTGAHQASPAHAMFLAGLLSGGKEDDITVA-AAWVCR 275
TPFS + Q +P TP + A Q P + +++ L E+++ V + V
Sbjct 393 TPFSAYSSSHGIHQRIPASTPTNTNPADQNPPCNTIYVGNLPPSTSEEELKVLFSTQVGY 452
Query 276 KDRCVYTENDG 286
K C T+ +G
Sbjct 453 KRLCFRTKGNG 463
> At3g63340_1
Length=750
Score = 31.2 bits (69), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 37/81 (45%), Gaps = 15/81 (18%)
Query 18 PADVAKEALTRAYH------------KARNFGSSTAIVGVLDGESGIFGLANLGDSSSLV 65
P D+ KEAL RA H + N GS+ I + DG+ +A++GDS +L+
Sbjct 176 PLDIMKEALLRAIHDIDVTFTKEASNRKLNSGSTATIALIADGQ---LMVASIGDSKALL 232
Query 66 LRRPRVRGSLGQLSLVKKVKE 86
+ +LVK +E
Sbjct 233 CSEKFETLEEARATLVKLYRE 253
> Hs22064245
Length=213
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 42/98 (42%), Gaps = 14/98 (14%)
Query 117 LVSIAKQEYDQGDGGRIGDEPSAIQSTSVPVQDGDLIILGTDGLFDNVFDFEAL------ 170
L+S + Y + GG +GD+P A T +++ DL G ++ DF+ L
Sbjct 112 LLSAGGRLYKEDWGGCVGDQPEAFH-TFPALKELDLAFNGIKTIYVKYGDFKLLEFLDLS 170
Query 171 -------ALTGLAVSPHEAQTLLGDASLATPPEDVARA 201
A+ L + PH LL L + P ++A A
Sbjct 171 FNSLTVEAICDLGILPHLRVLLLTGNGLTSLPPNLAVA 208
> Hs21361256
Length=299
Score = 30.8 bits (68), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 35/71 (49%), Gaps = 7/71 (9%)
Query 151 DLIILGTDGLFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPEDVARALALAAYWRS 210
D+++LGTDGL+D D E A +S +E D S T +A+AL L A
Sbjct 215 DVLVLGTDGLWDVTTDCEVAATVDRVLSAYEPN----DHSRYTA---LAQALVLGARGTP 267
Query 211 LDRDAQTPFSK 221
DR + P +K
Sbjct 268 RDRGWRLPNNK 278
> Hs5803219
Length=1064
Score = 30.0 bits (66), Expect = 7.0, Method: Composition-based stats.
Identities = 29/115 (25%), Positives = 44/115 (38%), Gaps = 25/115 (21%)
Query 148 QDGDLI-------ILGTDG-LFDNVFDFEALALTGLAVSPHEAQTLLGDASLATPPEDVA 199
+DGD I + GTDG +DN AL G+ + P +DV
Sbjct 105 RDGDFICPDYYEAVCGTDGKTYDNRC---ALCAENAKTGSQIGVKSEGECKSSNPEQDVC 161
Query 200 RALALAAYWRSLDRDAQTPFSKEARRCCNKQSLPVQTPDGSTGAHQASPAHAMFL 254
A PF ++ R C +++ PV PDG T ++ + +FL
Sbjct 162 SAF--------------RPFVRDGRLGCTRENDPVLGPDGKTHGNKCAMCAELFL 202
Lambda K H
0.314 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8375745028
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40