bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1974_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
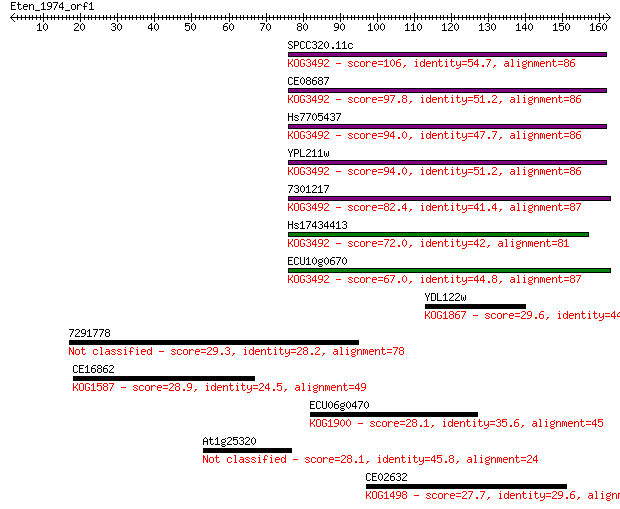
SPCC320.11c 106 2e-23
CE08687 97.8 8e-21
Hs7705437 94.0 1e-19
YPL211w 94.0 1e-19
7301217 82.4 4e-16
Hs17434413 72.0 5e-13
ECU10g0670 67.0 1e-11
YDL122w 29.6 2.5
7291778 29.3 3.1
CE16862 28.9 4.2
ECU06g0470 28.1 7.8
At1g25320 28.1 8.8
CE02632 27.7 9.0
> SPCC320.11c
Length=180
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 47/86 (54%), Positives = 63/86 (73%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLT +E K EKL++++G N+ HLI+ DPH FRL +DRV+++SE+ +K A V R
Sbjct 1 MRPLTHEETKTFFEKLAQYIGKNITHLIDRPDDPHCFRLQKDRVYYVSERAMKMATSVAR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
+NL+S+G C GK TK KFRL ITAL
Sbjct 61 QNLMSLGICFGKFTKTNKFRLHITAL 86
> CE08687
Length=180
Score = 97.8 bits (242), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 44/86 (51%), Positives = 61/86 (70%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLTE+E LV KL+ F+GDN+ LI+ + + FR H++RV++ SE L++ A C+ R
Sbjct 1 MRPLTEEETSLVFAKLASFIGDNVSMLIDRNDGDYCFRNHKERVYYCSENLMRQAACIAR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
+ LLS G C+GK TK +KF L ITAL
Sbjct 61 EPLLSFGTCLGKFTKSKKFHLQITAL 86
> Hs7705437
Length=180
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 41/86 (47%), Positives = 62/86 (72%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLTE+E +++ EK++K++G+NL L++ + FRLH DRV+++SEK++K A +
Sbjct 1 MRPLTEEETRVMFEKIAKYIGENLQLLVDRPDGTYCFRLHNDRVYYVSEKIMKLAANISG 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
L+S+G C GK TK KFRL +TAL
Sbjct 61 DKLVSLGTCFGKFTKTHKFRLHVTAL 86
> YPL211w
Length=181
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 44/86 (51%), Positives = 62/86 (72%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MR LTE+E K+V EKL+ ++G N+ L++N PHVFRL +DRV+++ + + K A V R
Sbjct 1 MRQLTEEETKVVFEKLAGYIGRNISFLVDNKELPHVFRLQKDRVYYVPDHVAKLATSVAR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
NL+S+G C+GK TK KFRL IT+L
Sbjct 61 PNLMSLGICLGKFTKTGKFRLHITSL 86
> 7301217
Length=180
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 58/87 (66%), Gaps = 0/87 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
M+ L+++ K++ E LSK++G N+ HLI+ + FR H+DRV+++SE++LK + C
Sbjct 1 MKRLSDERAKILFEHLSKYIGTNVKHLIDRPDGTYCFREHKDRVYYVSERILKLSECFGY 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITALH 162
K L+ VG C GK +K K + ITAL+
Sbjct 61 KQLVCVGTCFGKFSKTNKLKFHITALY 87
> Hs17434413
Length=133
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 34/81 (41%), Positives = 52/81 (64%), Gaps = 0/81 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLTE+E ++ EK+ K++ +NL L++ + F LH DRV ++SEK++K +
Sbjct 1 MRPLTEEETHVMFEKIVKYIRENLQLLLDRPDRTYCFWLHNDRVDYVSEKIVKLDANISG 60
Query 136 KNLLSVGQCIGKITKGRKFRL 156
L+S+G C GK+TK KFR
Sbjct 61 DKLVSLGTCFGKVTKTHKFRF 81
> ECU10g0670
Length=176
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/87 (44%), Positives = 53/87 (60%), Gaps = 3/87 (3%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MR L +E K VL+KL F+GDN+ ++ S + +F LH RV LSE++ A + R
Sbjct 1 MRDLKPEEEKEVLKKLRFFIGDNVHRIL--SGEDRLF-LHNQRVMILSERMRAATSMISR 57
Query 136 KNLLSVGQCIGKITKGRKFRLGITALH 162
KNL VG IGK TK FRLG+ +L+
Sbjct 58 KNLGMVGTVIGKFTKSGSFRLGVASLN 84
> YDL122w
Length=809
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 113 RLHRDRVFFLSEKLLKAAGCVPRKNLL 139
+ +RD+ +F + LLKA PRKN+L
Sbjct 213 KYYRDKPYFKTNSLLKAMSKSPRKNIL 239
> 7291778
Length=1726
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 22/78 (28%), Positives = 35/78 (44%), Gaps = 1/78 (1%)
Query 17 LSKLLKSFSLTYIEAQSNTLLFGSVEWYLLYSACTSTSPLHWENRIIAQKPLAWGWRLKM 76
LS+ +K S+ + AQ+ L + L TST PL+ + KP +R
Sbjct 1435 LSEQMKRASVATLPAQNQLRLITQLPANELVKVVTSTPPLN-SAEDVNSKPSMSTFRGST 1493
Query 77 RPLTEDEIKLVLEKLSKF 94
P T D++ LV + S+
Sbjct 1494 PPKTPDDLGLVTNRFSRL 1511
> CE16862
Length=643
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 12/49 (24%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 18 SKLLKSFSLTYIEAQSNTLLFGSVEWYLLYSACTSTSPLHWENRIIAQK 66
SK + + + Y +++ ++ F +LL + C S W+NR+ ++K
Sbjct 323 SKRMTAEFIVYCQSEIQSVAFARFHAHLLLAGCESGQICVWDNRLTSRK 371
> ECU06g0470
Length=962
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 4/45 (8%)
Query 82 DEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKL 126
D IK+ + L KFL + + +SSDP VF LH + ++K+
Sbjct 711 DVIKISVPFLEKFLKEKAM----SSSDPRVFELHWKYCVYRNDKV 751
> At1g25320
Length=702
Score = 28.1 bits (61), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 53 TSPLHWENRIIAQKPLAWGWRLKM 76
T+ LH +++ KPL+WG RLK+
Sbjct 487 TNALHGNPGMVSFKPLSWGVRLKI 510
> CE02632
Length=490
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 97 DNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPRKNLLSVGQCIGKITK 150
DNL HL + D +F++ +D + E LLKA + +K+ ++ + + I K
Sbjct 22 DNLAHLAAHGGDGRLFKMEQDYSKQVDEALLKARD-IAQKDAVAAVESLNNIEK 74
Lambda K H
0.324 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40