bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1975_orf4
Length=181
Score E
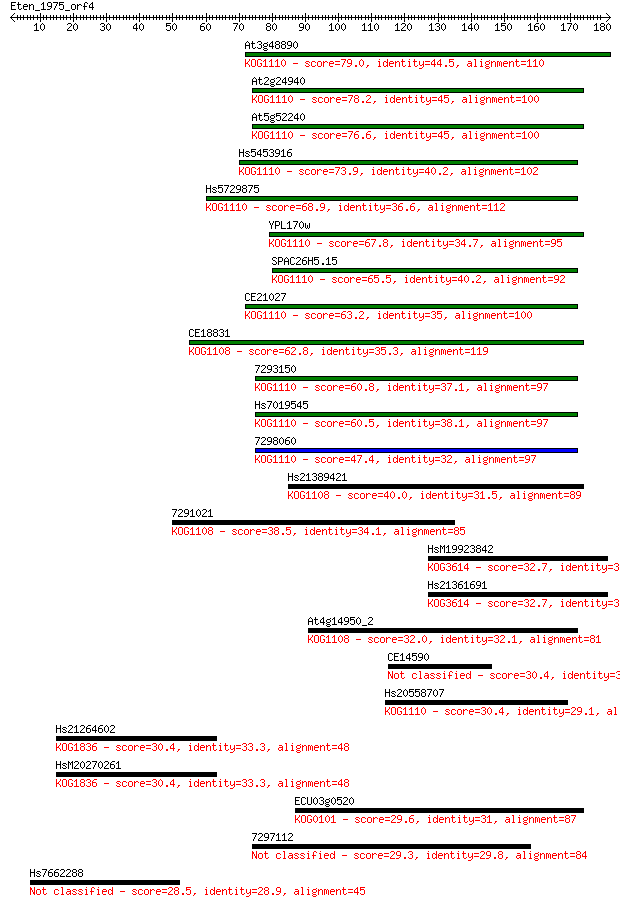
Sequences producing significant alignments: (Bits) Value
At3g48890 79.0 5e-15
At2g24940 78.2 8e-15
At5g52240 76.6 2e-14
Hs5453916 73.9 2e-13
Hs5729875 68.9 5e-12
YPL170w 67.8 1e-11
SPAC26H5.15 65.5 6e-11
CE21027 63.2 3e-10
CE18831 62.8 4e-10
7293150 60.8 1e-09
Hs7019545 60.5 2e-09
7298060 47.4 2e-05
Hs21389421 40.0 0.002
7291021 38.5 0.007
HsM19923842 32.7 0.36
Hs21361691 32.7 0.36
At4g14950_2 32.0 0.63
CE14590 30.4 1.8
Hs20558707 30.4 1.8
Hs21264602 30.4 2.1
HsM20270261 30.4 2.2
ECU03g0520 29.6 3.5
7297112 29.3 4.5
Hs7662288 28.5 8.5
> At3g48890
Length=233
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 49/111 (44%), Positives = 67/111 (60%), Gaps = 8/111 (7%)
Query 72 LPQMTLEALKSCDGEKSEK-LFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALAR 130
L ++T E LK DG S+K L +A+KG ++DVS + FYG G Y +FAGKDA+ ALA+
Sbjct 68 LGEITEEELKLYDGSDSKKPLLMAIKGQIYDVS-QSRMFYGPGGPYALFAGKDASRALAK 126
Query 131 MSFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVAVVDFEEKKDN 181
MSF+++ L S L E E+LQ W +F SKY V + +KKD
Sbjct 127 MSFEDQDLTGDIS---GLGAFELEALQDWEYKFMSKYVKVGTI---QKKDG 171
> At2g24940
Length=100
Score = 78.2 bits (191), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 45/101 (44%), Positives = 67/101 (66%), Gaps = 5/101 (4%)
Query 74 QMTLEALKSCDG-EKSEKLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMS 132
+ T E L +G ++S+ ++VA+KG VFDV+ GK+FYG G Y +FAGKDA+ AL +MS
Sbjct 2 EFTAEQLSQYNGTDESKPIYVAIKGRVFDVTT-GKSFYGSGGDYSMFAGKDASRALGKMS 60
Query 133 FDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVAVV 173
+E+ + SP + E LTE E +L W +F++KY +V V
Sbjct 61 KNEEDV-SP--SLEGLTEKEINTLNDWETKFEAKYPVVGRV 98
> At5g52240
Length=220
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/103 (43%), Positives = 63/103 (61%), Gaps = 9/103 (8%)
Query 74 QMTLEALKSCDGEKSEK-LFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMS 132
++T E LK DG +K L +A+K ++DV+ + FYG G Y +FAGKDA+ ALA+MS
Sbjct 74 EITEEELKQYDGSDPQKPLLMAIKHQIYDVT-QSRMFYGPGGPYALFAGKDASRALAKMS 132
Query 133 FDEKLLNSPPSTWE--SLTESEKESLQSWLQRFKSKYSLVAVV 173
F+EK L TW+ L E ++LQ W +F SKY+ V V
Sbjct 133 FEEKDL-----TWDVSGLGPFELDALQDWEYKFMSKYAKVGTV 170
> Hs5453916
Length=223
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 41/107 (38%), Positives = 57/107 (53%), Gaps = 6/107 (5%)
Query 70 TGLPQM-----TLEALKSCDGEKSEKLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDA 124
T LP+M +LE L+ DG ++ ++ +AV G VFDV+ G FYG G Y IFAG+DA
Sbjct 93 TSLPRMKKRDFSLEQLRQYDGSRNPRILLAVNGKVFDVT-KGSKFYGPAGPYGIFAGRDA 151
Query 125 TVALARMSFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVA 171
+ LA D+ L L + ES++ W +FK KY V
Sbjct 152 SRGLATFCLDKDALRDEYDDLSDLNAVQMESVREWEMQFKEKYDYVG 198
> Hs5729875
Length=195
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 62/118 (52%), Gaps = 7/118 (5%)
Query 60 AAAADVDVDTTG-LPQM-----TLEALKSCDGEKSEKLFVAVKGLVFDVSVNGKTFYGKN 113
AA+ D D D LP++ T L+ DG + ++ +A+ G VFDV+ G+ FYG
Sbjct 52 AASGDSDDDEPPPLPRLKRRDFTPAELRRFDGVQDPRILMAINGKVFDVT-KGRKFYGPE 110
Query 114 GSYHIFAGKDATVALARMSFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVA 171
G Y +FAG+DA+ LA D++ L LT +++E+L W +F KY V
Sbjct 111 GPYGVFAGRDASRGLATFCLDKEALKDEYDDLSDLTAAQQETLSDWESQFTFKYHHVG 168
> YPL170w
Length=152
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 56/98 (57%), Gaps = 4/98 (4%)
Query 79 ALKSCDGEKSEKLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMSFDEKLL 138
L +G EK+F+A++G V+D + G+ FYG +G Y FAG DA+ LA SFD ++
Sbjct 48 TLSKFNGHDDEKIFIAIRGKVYDCT-RGRQFYGPSGPYTNFAGHDASRGLALNSFDLDVI 106
Query 139 ---NSPPSTWESLTESEKESLQSWLQRFKSKYSLVAVV 173
+ P + LT+ + ++L W + F++KY + +
Sbjct 107 KDWDQPIDPLDDLTKEQIDALDEWQEHFENKYPCIGTL 144
> SPAC26H5.15
Length=166
Score = 65.5 bits (158), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 55/98 (56%), Gaps = 9/98 (9%)
Query 80 LKSCDGEKSEKLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMSFDEKLLN 139
LK +G K+ +F+A+KG V++V++ G FYG G Y FAG DA+ LA+ SFD++ +
Sbjct 48 LKEYNGSKNSLVFLAIKGTVYNVTM-GSKFYGPQGPYSAFAGHDASRGLAKNSFDDEFI- 105
Query 140 SPPSTWE------SLTESEKESLQSWLQRFKSKYSLVA 171
P S E L + E+++L W F KY V
Sbjct 106 -PDSDAEELDDCSDLNDEERQALNDWKAFFDQKYQAVG 142
> CE21027
Length=198
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 53/100 (53%), Gaps = 3/100 (3%)
Query 72 LPQMTLEALKSCDGEKSEKLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARM 131
+ MT+E L+ DG K+E + + G ++DV+ GK FYG +Y AG DAT AL M
Sbjct 62 MSDMTVEELRKYDGVKNEHILFGLNGTIYDVT-RGKGFYGPGKAYGTLAGHDATRALGTM 120
Query 132 SFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVA 171
D+ ++S ++ E+E+ W +FK KY V
Sbjct 121 --DQNAVSSEWDDHTGISADEQETANEWETQFKFKYLTVG 158
> CE18831
Length=326
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 59/120 (49%), Gaps = 5/120 (4%)
Query 55 AAAAAAAAADVDVDTTGLPQMTLEALKSCDGEKSEK-LFVAVKGLVFDVSVNGKTFYGKN 113
A A +VDV G T E L DG + K +++A+ G V++V K +YG
Sbjct 81 AEHIQAINPEVDVAAGGKHVFTPEQLHFFDGTRDSKPIYLAILGRVYNVD-GKKEYYGPG 139
Query 114 GSYHIFAGKDATVALARMSFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVAVV 173
SYH FAG+DAT A F E L +T L+ E S++ W+ + +Y LV VV
Sbjct 140 KSYHHFAGRDATRAFTTGDFQESGL---IATTHGLSHDELLSIRDWVSFYDKEYPLVGVV 196
> 7293150
Length=248
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 58/101 (57%), Gaps = 9/101 (8%)
Query 75 MTLEALKSCDGEKSE-KLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMSF 133
T++ L+ DG + + ++ VAV G V+DVS G+ FYG G Y FAG+DA+ LA S
Sbjct 83 FTVKELRQYDGTQPDGRVLVAVNGSVYDVS-KGRRFYGPGGPYATFAGRDASRNLATFS- 140
Query 134 DEKLLNSPPSTWESLTE---SEKESLQSWLQRFKSKYSLVA 171
+++ ++ L++ E +S++ W +FK KY LV
Sbjct 141 ---VVSIDKDEYDDLSDLSAVEMDSVREWEMQFKEKYELVG 178
> Hs7019545
Length=172
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/99 (37%), Positives = 59/99 (59%), Gaps = 6/99 (6%)
Query 75 MTLEALKSCDGEKSEK-LFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMSF 133
T E L GE+ ++ +++AVKG+VFDV+ +GK FYG+ Y+ GKD+T +A+MS
Sbjct 47 FTEEELARYGGEEEDQPIYLAVKGVVFDVT-SGKEFYGRGAPYNALTGKDSTRGVAKMSL 105
Query 134 DEKLLNSPPSTWESLTESEKESL-QSWLQRFKSKYSLVA 171
D L + LT E E+L + + + +K+KY +V
Sbjct 106 DPADLTHDTT---GLTAKELEALDEVFTKVYKAKYPIVG 141
> 7298060
Length=192
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 56/106 (52%), Gaps = 19/106 (17%)
Query 75 MTLEALKSCDGEKSE-KLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMSF 133
T+ L+ DG +++ ++ VA+ ++DVS +YG+NG +AG+D ++R+
Sbjct 73 FTVRELREYDGTRADGRILVAILFNIYDVS-RSVHYYGRNGVNPNYAGRD----ISRI-- 125
Query 134 DEKLLNSPPSTWES--------LTESEKESLQSWLQRFKSKYSLVA 171
L+NSP +S L+ ++ +L+ W QR+K KY V
Sbjct 126 ---LINSPEDLKDSEDFDDLSDLSRNQMNTLREWEQRYKMKYPFVG 168
> Hs21389421
Length=264
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 42/89 (47%), Gaps = 4/89 (4%)
Query 85 GEKSEKLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMSFDEKLLNSPPST 144
G L++A+ G V+DVS +G+ Y Y FAG+DA+ A E L S
Sbjct 49 GPGDPGLYLALLGRVYDVS-SGRRHYEPGSHYSGFAGRDASRAFVTGDCSEAGLVDDVS- 106
Query 145 WESLTESEKESLQSWLQRFKSKYSLVAVV 173
L+ +E +L +WL ++ Y V V
Sbjct 107 --DLSAAEMLTLHNWLSFYEKNYVCVGRV 133
> 7291021
Length=287
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 41/85 (48%), Gaps = 4/85 (4%)
Query 50 EQGAAAAAAAAAAADVDVDTTGLPQMTLEALKSCDGEKSEKLFVAVKGLVFDVSVNGKTF 109
+ G A+ A A D+ T P E K E+ L++A+ G VFDVS G
Sbjct 43 QAGQDASIPLAFQAGDDIGTLFTPA---ELAKFNGEEEGRPLYLALLGSVFDVS-RGIKH 98
Query 110 YGKNGSYHIFAGKDATVALARMSFD 134
YG SY+ F G+DA+V+ F+
Sbjct 99 YGSGCSYNFFVGRDASVSFISGDFE 123
> HsM19923842
Length=1095
Score = 32.7 bits (73), Expect = 0.36, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 24/54 (44%), Gaps = 0/54 (0%)
Query 127 ALARMSFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVAVVDFEEKKD 180
AL + EKL+ P T L E E ES WL+ L+ V+ EE D
Sbjct 339 ALTSSAVKEKLVRFLPRTVSRLPEEETESWIKWLKEILECSHLLTVIKMEEAGD 392
> Hs21361691
Length=1104
Score = 32.7 bits (73), Expect = 0.36, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 24/54 (44%), Gaps = 0/54 (0%)
Query 127 ALARMSFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYSLVAVVDFEEKKD 180
AL + EKL+ P T L E E ES WL+ L+ V+ EE D
Sbjct 348 ALTSSAVKEKLVRFLPRTVSRLPEEETESWIKWLKEILECSHLLTVIKMEEAGD 401
> At4g14950_2
Length=177
Score = 32.0 bits (71), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 41/94 (43%), Gaps = 17/94 (18%)
Query 91 LFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGK-------------DATVALARMSFDEKL 137
+ + + G VFDV+ GK YG G Y+ FAG+ DA+ A +F
Sbjct 21 ILLGILGSVFDVT-KGKFHYGSGGGYNHFAGRFVLLKIMWRDSHIDASRAFVSGNFTGDG 79
Query 138 LNSPPSTWESLTESEKESLQSWLQRFKSKYSLVA 171
L + + L+ SE +S+ W + Y+ V
Sbjct 80 L---TDSLQGLSSSEVKSIVDWRGFYSRTYTPVG 110
> CE14590
Length=363
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 115 SYHIFAGKDATVALARMSFDEKLLNSPPSTW 145
SY I+ G + L + FDE+L SPP +
Sbjct 38 SYQIYGGLPIALGLTELLFDERLQESPPHNF 68
> Hs20558707
Length=215
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 25/55 (45%), Gaps = 0/55 (0%)
Query 114 GSYHIFAGKDATVALARMSFDEKLLNSPPSTWESLTESEKESLQSWLQRFKSKYS 168
G + IFA +D + A D+ L L +++E+L W +F KYS
Sbjct 60 GPHRIFAERDTSRGPATFCLDKGALQDKHDDLSDLPPAQEETLSDWASQFTFKYS 114
> Hs21264602
Length=3695
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 22/48 (45%), Gaps = 0/48 (0%)
Query 15 LPKSPAAAPAAPRCCSCRIRNSSSASPTKMEKAPCEQGAAAAAAAAAA 62
LP+ A P P C C R + + SP M ++ GA A+ AA
Sbjct 3648 LPEPMAVQPWPPAYCGCMRRLAVNRSPVAMTRSVEVHGAVGASGCPAA 3695
> HsM20270261
Length=3695
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 22/48 (45%), Gaps = 0/48 (0%)
Query 15 LPKSPAAAPAAPRCCSCRIRNSSSASPTKMEKAPCEQGAAAAAAAAAA 62
LP+ A P P C C R + + SP M ++ GA A+ AA
Sbjct 3648 LPEPMAVQPWPPAYCGCMRRLAVNRSPVAMTRSVEVHGAVGASGCPAA 3695
> ECU03g0520
Length=683
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 38/89 (42%), Gaps = 2/89 (2%)
Query 87 KSEKLFVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATVALARMSFDEKLLNSPPSTWE 146
K + L + G FDVS+ F G GS I D L FD L+N S +
Sbjct 225 KEDVLVFDLGGGTFDVSLLDFEFNGAAGSLGIVKAIDGDTFLGGQDFDNLLINYCISEFL 284
Query 147 SLTESEKES--LQSWLQRFKSKYSLVAVV 173
S K+S +S L R +++ + V V
Sbjct 285 KKNSSIKQSDLKESALLRLRAECTRVKAV 313
> 7297112
Length=2168
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 25/92 (27%), Positives = 44/92 (47%), Gaps = 13/92 (14%)
Query 74 QMTLEALKSCDGEKSEKL-------FVAVKGLVFDVSVNGKTFYGKNGSYHIFAGKDATV 126
Q TL L+S G+ ++L ++ +GL+ D+ + G NG Y + AG
Sbjct 1904 QWTLSRLQSRYGKFGQELVAWRRTLYLLSQGLI-DLPADS----GSNGGYVVDAGSTNAT 1958
Query 127 ALARMSFDEKLLNSPPSTW-ESLTESEKESLQ 157
A+ + F + + PP T+ ES E ++ L+
Sbjct 1959 AVYKPIFSKDRGDQPPHTYDESFVEGDEPGLE 1990
> Hs7662288
Length=967
Score = 28.5 bits (62), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 13/46 (28%), Positives = 26/46 (56%), Gaps = 5/46 (10%)
Query 7 LSSKKGPFLPKSPAAAPAAPRCCS-CRIRNSSSASPTKMEKAPCEQ 51
+ +K P++P +P + PA+P+C + +++ SP K P +Q
Sbjct 550 VKDRKAPWIPPNPTSPPASPKCAAWLKVKT----SPRDATKEPLQQ 591
Lambda K H
0.313 0.126 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2842629034
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40