bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1977_orf2
Length=265
Score E
Sequences producing significant alignments: (Bits) Value
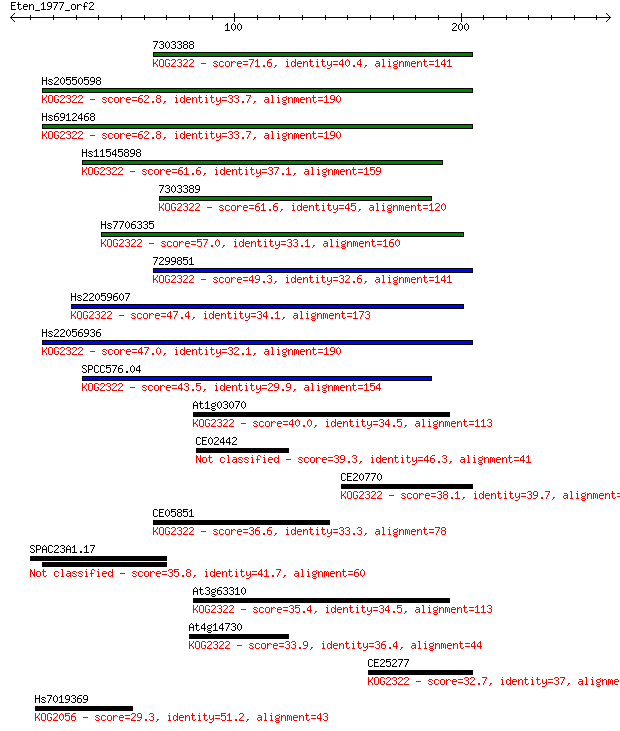
7303388 71.6 1e-12
Hs20550598 62.8 7e-10
Hs6912468 62.8 8e-10
Hs11545898 61.6 2e-09
7303389 61.6 2e-09
Hs7706335 57.0 4e-08
7299851 49.3 9e-06
Hs22059607 47.4 3e-05
Hs22056936 47.0 4e-05
SPCC576.04 43.5 5e-04
At1g03070 40.0 0.005
CE02442 39.3 0.008
CE20770 38.1 0.020
CE05851 36.6 0.063
SPAC23A1.17 35.8 0.094
At3g63310 35.4 0.12
At4g14730 33.9 0.34
CE25277 32.7 0.83
Hs7019369 29.3 8.0
> 7303388
Length=324
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 57/176 (32%), Positives = 79/176 (44%), Gaps = 35/176 (19%)
Query 64 GSPPAPSDDTKITDHISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNY 123
G+ P K IR F+RKV IL Q++ TFG ++F + +TF N
Sbjct 90 GAYDDPESQPKNFSFDDQSIRRGFIRKVYLILMGQLIVTFGAVALFVYHEGTKTFARNNM 149
Query 124 WLAIVA-----------AVCGLV----------LQLASSWESF--------------LIA 148
WL VA A C V L L ++ +SF L+A
Sbjct 150 WLFWVALGVMLVTMLSMACCESVRRQTPTNFIFLGLFTAAQSFLMGVSATKYAPKEVLMA 209
Query 149 IGSTFVVVVGLMLFTCQTKYDFTGCGTYLFVAVLCLMIFGILSIFFHNKIVHLVYS 204
+G T V + L +F QTKYDFT G L ++ +IFGI++IF KI+ LVY+
Sbjct 210 VGITAAVCLALTIFALQTKYDFTMMGGILIACMVVFLIFGIVAIFVKGKIITLVYA 265
> Hs20550598
Length=316
Score = 62.8 bits (151), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 64/229 (27%), Positives = 99/229 (43%), Gaps = 45/229 (19%)
Query 15 PPYPAACGGPYGGMAPPYPPVSPAYPAAGGGPSPVYGKPAGSTPGSMEMGSPPAPSDDTK 74
P Y A G G A +PP A P PS Y P+ S+ S + G P +
Sbjct 36 PSYEEATSGE-GMKAGAFPPAPTAVPL---HPSWAYVDPSSSS--SYDNGFPTGDHELFT 89
Query 75 ITDHISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---YWLA----- 126
++R FVRKV IL +Q+L T + ++F F ++ ++ N YW +
Sbjct 90 TFSWDDQKVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPVKDYVQANPGWYWASYAVFF 149
Query 127 ---IVAAVC---------GLVL-------------QLASSWE--SFLIAIGSTFVVVVGL 159
+ A C L+L L+S + S L+ +G T +V + +
Sbjct 150 ATYLTLACCSGPRRHFPWNLILLTVFTLSMAYLTGMLSSYYNTTSVLLCLGITALVCLSV 209
Query 160 MLFTCQTKYDFTGCGTYLFVAVLCL----MIFGILSIFFHNKIVHLVYS 204
+F+ QTK+DFT C LFV ++ L +I IL F + +H VY+
Sbjct 210 TVFSFQTKFDFTSCQGVLFVLLMTLFFSGLILAILLPFQYVPWLHAVYA 258
> Hs6912468
Length=316
Score = 62.8 bits (151), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 64/229 (27%), Positives = 100/229 (43%), Gaps = 45/229 (19%)
Query 15 PPYPAACGGPYGGMAPPYPPVSPAYPAAGGGPSPVYGKPAGSTPGSMEMGSPPAPSDDTK 74
P Y A G G A +PP A P PS Y P+ S+ S + G PP +
Sbjct 36 PSYEEATSGE-GMKAGAFPPAPTAVPL---HPSWAYVDPSSSS--SYDNGFPPETMSSSP 89
Query 75 ITDHISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---YWLA----- 126
++ ++ ++R FVRKV IL +Q+L T + ++F F + YW +
Sbjct 90 LSAGMTKKVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPCQGLCSGQPGWYWASYAVFF 149
Query 127 ---IVAAVC---------GLVL-------------QLASSWE--SFLIAIGSTFVVVVGL 159
+ A C L+L L+S + S L+ +G T +V + +
Sbjct 150 ATYLTLACCSGPRRHFPWNLILLTVFTLSMAYLTGMLSSYYNTTSVLLCLGITALVCLSV 209
Query 160 MLFTCQTKYDFTGCGTYLFVAVLCL----MIFGILSIFFHNKIVHLVYS 204
+F+ QTK+DFT C LFV ++ L +I IL F + +H VY+
Sbjct 210 TVFSFQTKFDFTSCQGVLFVLLMTLFFSGLILAILLPFQYVPWLHAVYA 258
> Hs11545898
Length=311
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 59/209 (28%), Positives = 93/209 (44%), Gaps = 50/209 (23%)
Query 33 PPVSPA-YPAAGGGPSPVYGKPAGST--------------PGSMEMGSPPAPSDDTKITD 77
P V P YPA G P P YG PAG PG G A SD +
Sbjct 29 PSVLPGGYPAYPGYPQPGYGHPAGYPQPMPPTHPMPMNYGPGHGYDGEERAVSDSFGPGE 88
Query 78 HISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---YWLA-------- 126
++RH F+RKV I++VQ+L T I ++F FV + F+ +N Y+++
Sbjct 89 WDDRKVRHTFIRKVYSIISVQLLITVAIIAIFTFVEPVSAFVRRNVAVYYVSYAVFVVTY 148
Query 127 IVAAVC---------GLVL-------------QLASSWES--FLIAIGSTFVVVVGLMLF 162
++ A C ++L ++S +++ +IA+ T VV + + +F
Sbjct 149 LILACCQGPRRRFPWNIILLTLFTFAMGFMTGTISSMYQTKAVIIAMIITAVVSISVTIF 208
Query 163 TCQTKYDFTGCGTYLFVAVLCLMIFGILS 191
QTK DFT C V + L++ GI++
Sbjct 209 CFQTKVDFTSCTGLFCVLGIVLLVTGIVT 237
> 7303389
Length=239
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 54/155 (34%), Positives = 72/155 (46%), Gaps = 40/155 (25%)
Query 67 PAPSDDTKITDHISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---Y 123
A +D + D S IR F+RKV IL Q+L TFG SVF F + ++ +N +
Sbjct 10 DAEADKSFAFDDQS--IRKGFIRKVYLILMCQLLITFGFVSVFTFSKASQEWVQKNPALF 67
Query 124 WLA----IVAAVCG--------------LVLQLASSWESFLI--------------AIGS 151
W+A IV +C + L L + ESFL+ A+G
Sbjct 68 WIALAVLIVTMICMACCESVRRKTPLNFIFLFLFTVAESFLLGMVAGQFEADEVLMAVGI 127
Query 152 TFVVVVGLMLFTCQTKYDFTGCGTYLFVAVLCLMI 186
T V +GL LF QTKYDFT CG V V CL++
Sbjct 128 TAAVALGLTLFALQTKYDFTMCGG---VLVACLVV 159
> Hs7706335
Length=258
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 53/192 (27%), Positives = 85/192 (44%), Gaps = 43/192 (22%)
Query 41 AAGGGPSPVYGKPAGSTPGSMEMGSPPAPSDDTKITDHISVQIRHAFVRKVLGILAVQIL 100
A P P Y P S GS A + +V IR AF+RKV IL++Q+L
Sbjct 19 AIMADPDPRY--PRSSIEDDFNYGSSVASA---------TVHIRMAFLRKVYSILSLQVL 67
Query 101 FTFGIASVFGFVPTLRTFLLQNYWLAIVAAV--CGLVLQLA----------------SSW 142
T ++VF + ++RTF+ ++ L ++ A+ GL+ L +
Sbjct 68 LTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALTLNRHKYPLNLYLLFGFTLL 127
Query 143 ESFLIAIGSTF--------------VVVVGLMLFTCQTKYDFTGCGTYLFVAVLCLMIFG 188
E+ +A+ TF V GL ++T Q+K DF+ G LF + L + G
Sbjct 128 EALTVAVVVTFYDVYIILQAFILTTTVFFGLTVYTLQSKKDFSKFGAGLFALLWILCLSG 187
Query 189 ILSIFFHNKIVH 200
L FF+++I+
Sbjct 188 FLKFFFYSEIME 199
> 7299851
Length=264
Score = 49.3 bits (116), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 46/176 (26%), Positives = 73/176 (41%), Gaps = 35/176 (19%)
Query 64 GSPPAPSDDTKITDHISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNY 123
GS D K S IR F+RKV IL Q++ + + +R + ++
Sbjct 31 GSEIGQDDPDKGLGFCSASIRRGFIRKVYLILLAQLITSLVVIVSLTADKRVRLMVAEST 90
Query 124 WLAIVAAV----------CG-----------LVLQLASSWESFLIAIGS----------- 151
W+ +VA + C + L + ESFL+ + +
Sbjct 91 WIFVVAILIVVFSLVALGCNEDLRRQTPANFIFLSAFTIAESFLLGVAACRYAPMEIFMA 150
Query 152 ---TFVVVVGLMLFTCQTKYDFTGCGTYLFVAVLCLMIFGILSIFFHNKIVHLVYS 204
T V +GL LF QT+YDFT G L ++ L+ FGI++IF +V +Y+
Sbjct 151 VLITASVCLGLTLFALQTRYDFTVMGGLLVSCLIILLFFGIVTIFVGGHMVTTIYA 206
> Hs22059607
Length=285
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 59/230 (25%), Positives = 92/230 (40%), Gaps = 61/230 (26%)
Query 28 MAPPYPPVSPAYPAAGGGPSPVYGKPAGS-------------TPGSMEMGSPPAPSDDTK 74
MA P P YP + YG S T M P APS
Sbjct 1 MADP----DPRYPRSSIEDDFNYGSSVASATVHIRMVLQRDCTFSKQYMRIPSAPSATLN 56
Query 75 ITD--HIS-------VQIRH---AFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN 122
I H+ V I + F+RKV IL++Q+L T ++VF + ++RTF+ ++
Sbjct 57 IIRFFHLRQSGRSGMVNIFYKGPTFLRKVYSILSLQVLLTTVTSTVFLYFESVRTFVHES 116
Query 123 YWLAIVAAV--CGLVLQLA----------------SSWESFLIAIGSTF----------- 153
L ++ A+ GL+ L + E+ +A+ TF
Sbjct 117 PALILLFALGSLGLIFALILNRHKYPLNLYLLFGFTLLEALTVAVVVTFYDVYIILQAFI 176
Query 154 ---VVVVGLMLFTCQTKYDFTGCGTYLFVAVLCLMIFGILSIFFHNKIVH 200
V GL ++T Q+K DF+ G LF + L + G L FF+++I+
Sbjct 177 LTTTVFFGLTVYTLQSKKDFSKFGAGLFALLWILCLSGFLKFFFYSEIME 226
> Hs22056936
Length=349
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 61/256 (23%), Positives = 94/256 (36%), Gaps = 72/256 (28%)
Query 15 PPYPAACGGPYGGMAPPYPPVSPAYPAAGGGPSPVYGKPAGSTPGSMEMGSPPAPSDDTK 74
P Y A G G A +PP A P PS Y P+ S+ S + G P +
Sbjct 40 PSYEEATSGE-GMKAGAFPPAPTAVPL---HPSWAYVDPSSSS--SYDNGFPTGDHELFT 93
Query 75 ITDHISVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN---YWLA----- 126
++R FVRKV IL +Q+L T + ++F F ++ ++ N YW +
Sbjct 94 TFSWDDQKVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPVKDYVQANPGWYWASYAVFF 153
Query 127 ---IVAAVC-------GLVLQLAS-------------------SWESFLIAI-------- 149
+ A C GL L W L+ +
Sbjct 154 ATYLTLACCSGPRYANGLRRNLWEYEQDHENSGPSPASHQRHFPWNLILLTVFTLSMAYL 213
Query 150 ---------GSTFVVVVGLMLFTC--------QTKYDFTGCGTYLFVAVLCL----MIFG 188
++ ++ +G+ C QTK+DFT C LFV ++ L +I
Sbjct 214 TGMLSSYYNTTSVLLCLGITALVCLSVTVFSFQTKFDFTSCQGVLFVLLMTLFFSGLILA 273
Query 189 ILSIFFHNKIVHLVYS 204
IL F + +H VY+
Sbjct 274 ILLPFQYVPWLHAVYA 289
> SPCC576.04
Length=266
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 46/188 (24%), Positives = 76/188 (40%), Gaps = 41/188 (21%)
Query 33 PPVSPAYPAAGGGPSPV--YGKPAGSTPGSMEMGSPPAPSDDTKITDHISVQIRHAFVRK 90
P P+ P P Y + A P + + ++ K IR AF+RK
Sbjct 9 PQDDPSVPNLAQAPPAYSEYNESATENPAVDQFKNTTPVAECAK-------SIRMAFLRK 61
Query 91 VLGILAVQILFTFGIASVFGFVPTLRT-------FLLQNYWLAIV--------------- 128
V IL Q+ T +F P FL+ N+++++V
Sbjct 62 VYAILTAQLFVTSLFGGIFYLHPAFSFWVQMHPWFLILNFFISLVVLFGLIMKPYSYPRN 121
Query 129 -------AAVCGLVLQLASSWESFLIAIGSTFV---VVVGLMLFTCQTKYDFTGCGTYLF 178
A+ GL L A ++ S I + + F+ V V L FT Q+K+DF+ G +L+
Sbjct 122 YIFLFLFTALEGLTLGTAITFFSARIILEAVFITLGVFVALTAFTFQSKWDFSRLGGFLY 181
Query 179 VAVLCLMI 186
V++ L++
Sbjct 182 VSLWSLIL 189
> At1g03070
Length=247
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 39/151 (25%), Positives = 65/151 (43%), Gaps = 38/151 (25%)
Query 82 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQN-----YWLAIVAA------ 130
++R F+RKV I+A Q+L T +AS FV + F W+ ++
Sbjct 33 ELRWGFIRKVYSIIAFQLLATIAVASTVVFVRPIAVFFATTSAGLALWIVLIITPLIVMC 92
Query 131 ----------VCGLVLQLASSWESFLIAIGSTF--------------VVVVGLMLFT--- 163
V L+L + + +F + + F VVV+ L ++T
Sbjct 93 PLYYYHQKHPVNYLLLGIFTVALAFAVGLTCAFTSGKVILEAAILTTVVVLSLTVYTFWA 152
Query 164 CQTKYDFTGCGTYLFVAVLCLMIFGILSIFF 194
+ YDF G +LF A++ LM+F ++ IFF
Sbjct 153 AKKGYDFNFLGPFLFGALIVLMVFALIQIFF 183
> CE02442
Length=130
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 83 IRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNY 123
IR AF+RKVLGI+ Q+LFT GI + +P L + Y
Sbjct 69 IRIAFLRKVLGIVGFQLLFTIGICAAIYNIPNSNQLLQKQY 109
> CE20770
Length=295
Score = 38.1 bits (87), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 33/62 (53%), Gaps = 8/62 (12%)
Query 147 IAIGSTFVVVVGLMLFTCQTKYDFTGCGTYLFVAVLCLMIFG----ILSIFFHNKIVHLV 202
I IG TF +V+ QTK+D T Y+ + +C M FG I S+FF K + +V
Sbjct 182 ICIGCTFSIVI----VASQTKFDLTAHMGYILIISMCFMFFGLVVVICSMFFKIKFLMMV 237
Query 203 YS 204
Y+
Sbjct 238 YA 239
> CE05851
Length=244
Score = 36.6 bits (83), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 43/84 (51%), Gaps = 10/84 (11%)
Query 64 GSPPAPSD-DTKITDHISVQ-IRHAFVRKVLGILAVQILFTFGIASVFGFVPTL----RT 117
G+ A D D K H S Q +R AFVRKV ++ + F I + F +P + +
Sbjct 6 GATTAQDDPDGKYNLHFSSQTVRAAFVRKVFMLVTIM----FAITAAFCVIPMVSEPFQD 61
Query 118 FLLQNYWLAIVAAVCGLVLQLASS 141
++ N+W+ +A + LV+ +A S
Sbjct 62 WVKNNFWVYFIAIIVFLVVAIALS 85
> SPAC23A1.17
Length=1611
Score = 35.8 bits (81), Expect = 0.094, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 28/67 (41%), Gaps = 8/67 (11%)
Query 10 AQYGGPPYPAACGGPY----GGMAPPYPPVS---PAYPAAGGGPSPVYGKPAGSTPGSME 62
A G PP PA G P APP P S P PA G P PV + P +
Sbjct 1078 APSGAPPVPAPSGIPPVPKPSVAAPPVPKPSVAVPPVPAPSGAP-PVPKPSVAAPPVPVP 1136
Query 63 MGSPPAP 69
G+PP P
Sbjct 1137 SGAPPVP 1143
Score = 31.6 bits (70), Expect = 1.7, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 23/58 (39%), Gaps = 10/58 (17%)
Query 15 PPYPAACGGPYGGMAPPYPPVS---PAYPAAGGGPSPVYGKPAGSTPGSMEMGSPPAP 69
PP PA G APP P S P P G P PV + P G+PP P
Sbjct 1112 PPVPAPSG------APPVPKPSVAAPPVPVPSGAP-PVPKPSVAAPPVPAPSGAPPVP 1162
> At3g63310
Length=239
Score = 35.4 bits (80), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 39/152 (25%), Positives = 63/152 (41%), Gaps = 40/152 (26%)
Query 82 QIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQ-------------------- 121
++R +F+RKV I+++Q+L T +A+ V ++ F
Sbjct 24 ELRWSFIRKVYSIISIQLLVTIAVAATVVKVHSISVFFTTTTAGFALYILLILTPLIVMC 83
Query 122 -----------NYWL-----AIVAAVCGLVLQLASSWESFLIAIGSTFVVVVGLMLFT-- 163
NY L +A GL S + L ++ T VVV+ L L+T
Sbjct 84 PLYYYHQKHPVNYLLLGIFTVALAFAVGLTCAFTSG-KVILESVILTAVVVISLTLYTFW 142
Query 164 -CQTKYDFTGCGTYLFVAVLCLMIFGILSIFF 194
+ +DF G +LF AV+ LM+F + I F
Sbjct 143 AAKRGHDFNFLGPFLFGAVIVLMVFSFIQILF 174
> At4g14730
Length=100
Score = 33.9 bits (76), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 80 SVQIRHAFVRKVLGILAVQILFTFGIASVFGFVPTLRTFLLQNY 123
S ++R AF+RK+ IL++Q+L T G+++V FV + F+ + +
Sbjct 21 SSELRWAFIRKLYSILSLQLLVTVGVSAVVYFVRPIPEFITETH 64
> CE25277
Length=133
Score = 32.7 bits (73), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 31/50 (62%), Gaps = 4/50 (8%)
Query 159 LMLFTCQTKYDFTGCGTYLFVAVLCLMIFGILS----IFFHNKIVHLVYS 204
++LF TK D T C F+ +CLM+FG+++ IF + + +++VY+
Sbjct 28 IILFAATTKKDLTSCLGVAFILGICLMLFGLMACIFCIFLNWQFLYIVYA 77
> Hs7019369
Length=551
Score = 29.3 bits (64), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 22/45 (48%), Positives = 24/45 (53%), Gaps = 3/45 (6%)
Query 12 YGGPPYPAACGGPYGGMAPPYPPVSPAYPAAGGGPS--PVYGKPA 54
+GGPP P A P+GG AP P PAA GPS P G PA
Sbjct 271 WGGPPVPPAAD-PWGGPAPTPASGDPWRPAAPAGPSVDPWGGTPA 314
Lambda K H
0.326 0.143 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5649423594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40