bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2055_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
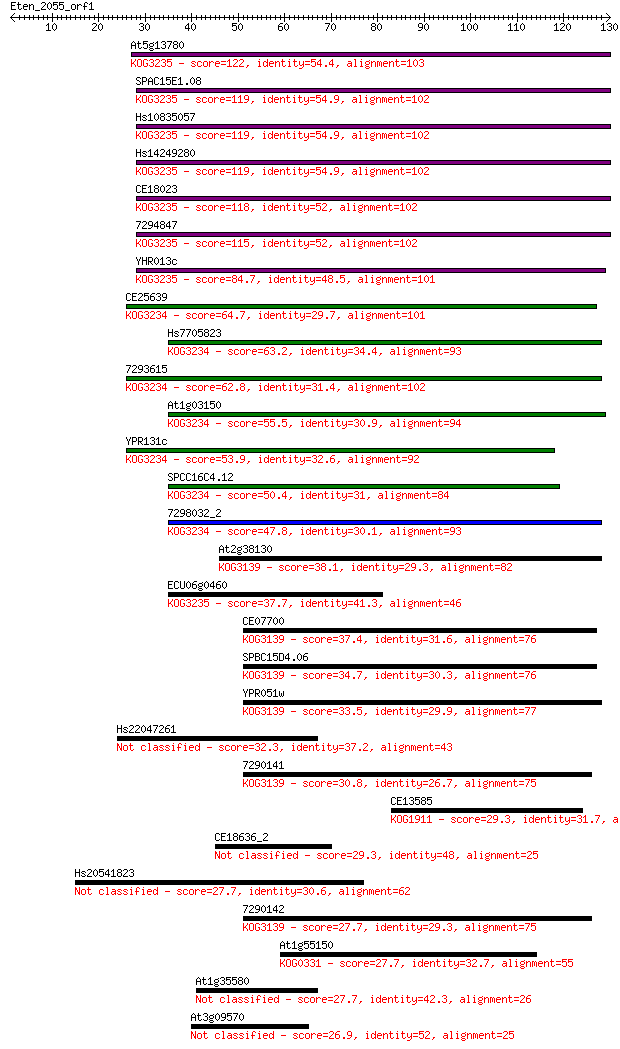
At5g13780 122 1e-28
SPAC15E1.08 119 1e-27
Hs10835057 119 1e-27
Hs14249280 119 1e-27
CE18023 118 3e-27
7294847 115 2e-26
YHR013c 84.7 4e-17
CE25639 64.7 5e-11
Hs7705823 63.2 1e-10
7293615 62.8 1e-10
At1g03150 55.5 3e-08
YPR131c 53.9 7e-08
SPCC16C4.12 50.4 8e-07
7298032_2 47.8 5e-06
At2g38130 38.1 0.004
ECU06g0460 37.7 0.005
CE07700 37.4 0.007
SPBC15D4.06 34.7 0.046
YPR051w 33.5 0.11
Hs22047261 32.3 0.20
7290141 30.8 0.67
CE13585 29.3 1.7
CE18636_2 29.3 1.9
Hs20541823 27.7 5.4
7290142 27.7 5.7
At1g55150 27.7 5.9
At1g35580 27.7 5.9
At3g09570 26.9 9.0
> At5g13780
Length=192
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 56/103 (54%), Positives = 77/103 (74%), Gaps = 3/103 (2%)
Query 27 LVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLE 86
+R + V DL MQ N + LPENY MKY+ +H LSWPQL VA+D G++VGYVLAK+E
Sbjct 3 CIRRATVDDLLAMQACNLMCLPENYQMKYYLYHILSWPQLLYVAEDYNGRIVGYVLAKME 62
Query 87 DENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+E+ + HGH+TS+AV+R +RKLGLA+KLM +Q AM++V+
Sbjct 63 EESNEC---HGHITSLAVLRTHRKLGLATKLMTAAQAAMEQVY 102
> SPAC15E1.08
Length=177
Score = 119 bits (299), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 76/102 (74%), Gaps = 1/102 (0%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+RP+ + DL MQ+ N NLPENY +KY+ +H++SWP L VA D +G++VGYVLAK+E+
Sbjct 3 IRPARISDLTGMQNCNLHNLPENYQLKYYLYHAISWPMLSYVATDPKGRVVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
E + PHGH+TSV+VMR R LGLA +LM SQRAM +V+
Sbjct 63 EPKD-GIPHGHITSVSVMRSYRHLGLAKRLMVQSQRAMVEVY 103
> Hs10835057
Length=235
Score = 119 bits (299), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 73/102 (71%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+F+H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNARPEDLMNMQHCNLLCLPENYQMKYYFYHGLSWPQLSYIAEDENGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 DPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> Hs14249280
Length=229
Score = 119 bits (298), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 72/102 (70%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+ +H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNAQPDDLMNMQHCNLLCLPENYQMKYYLYHGLSWPQLSYIAEDEDGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
E PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 EPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> CE18023
Length=182
Score = 118 bits (295), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 77/102 (75%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + V DL MQ+ N + LPENY MKY+F+H+LSWPQL +A+D +G +VGYVLAK+E+
Sbjct 3 IRCARVDDLMSMQNANLMCLPENYQMKYYFYHALSWPQLSYIAEDHKGNVVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ ++PHGH+TS+AV R R+LGLA+K+M + RAM + +
Sbjct 63 D--PGEEPHGHITSLAVKRSYRRLGLANKMMDQTARAMVETY 102
> 7294847
Length=196
Score = 115 bits (287), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 70/102 (68%), Gaps = 0/102 (0%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+F+H L+WPQL VA D +G +VGYVLAK+E+
Sbjct 3 IRCAKPEDLMTMQHCNLLCLPENYQMKYYFYHGLTWPQLSYVAVDDKGAIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ HGH+TS+AV R R+LGLA KLM + +AM + F
Sbjct 63 PEPNEESRHGHITSLAVKRSYRRLGLAQKLMNQASQAMVECF 104
> YHR013c
Length=238
Score = 84.7 bits (208), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 75/140 (53%), Gaps = 39/140 (27%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVA----------------- 70
+R + + D+ MQ+ N NLPENY+MKY+ +H LSWP+ VA
Sbjct 5 IRRATINDIICMQNANLHNLPENYMMKYYMYHILSWPEASFVATTTTLDCEDSDEQDEND 64
Query 71 ---------QDGQG------------KLVGYVLAKLEDE-NRQFQKPHGHVTSVAVMRQN 108
DG+ KLVGYVL K+ D+ ++Q + P+GH+TS++VMR
Sbjct 65 KLELTLDGTNDGRTIKLDPTYLAPGEKLVGYVLVKMNDDPDQQNEPPNGHITSLSVMRTY 124
Query 109 RKLGLASKLMRLSQRAMQKV 128
R++G+A LMR + A+++V
Sbjct 125 RRMGIAENLMRQALFALREV 144
> CE25639
Length=173
Score = 64.7 bits (156), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 60/101 (59%), Gaps = 3/101 (2%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKL 85
+ +RP DV D+F+ ++N E Y +++ + +++P+ VA+ G+++ YV+ K+
Sbjct 2 TTLRPFDVMDMFKFNNVNLDINTETYGFQFYLHYMMNYPEYYQVAEHPNGEIMAYVMGKI 61
Query 86 EDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQ 126
E + + HGHVT+++V R+LGLA+ +M +R +
Sbjct 62 EGRDTNW---HGHVTALSVAPNFRRLGLAAYMMEFLERTSE 99
> Hs7705823
Length=178
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 56/93 (60%), Gaps = 1/93 (1%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DLF ++N L E Y + ++ + WP+ VA+ G+L+GY++ K E + ++
Sbjct 11 DLFRFNNINLDPLTETYGIPFYLQYLAHWPEYFIVAEAPGGELMGYIMGKAEGSVAR-EE 69
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQK 127
HGHVT+++V + R+LGLA+KLM L + ++
Sbjct 70 WHGHVTALSVAPEFRRLGLAAKLMELLEEISER 102
> 7293615
Length=175
Score = 62.8 bits (151), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 56/102 (54%), Gaps = 3/102 (2%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKL 85
+ +RP DLF+ ++NF L E Y + ++ + WP+ +A+ G+++GY++ K+
Sbjct 2 TTLRPFTCDDLFKFNNVNFDPLTETYGLSFYTQYLAKWPEYFQLAESPSGQIMGYIMGKV 61
Query 86 EDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQK 127
E HGHVT++ V R+LGLA+ LM + +K
Sbjct 62 EG---HLDNWHGHVTALTVSPDYRRLGLAALLMSFLEDISEK 100
> At1g03150
Length=174
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 49/94 (52%), Gaps = 3/94 (3%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DL +N +L E + M ++ + WP VA+ +++GY++ K+E + +
Sbjct 11 DLLRFTSVNLDHLTETFNMSFYMTYLARWPDYFHVAEGPGNRVMGYIMGKVEGQGESW-- 68
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKV 128
HGHVT+V V + R+ LA KLM L + K+
Sbjct 69 -HGHVTAVTVSPEYRRQQLAKKLMNLLEDISDKI 101
> YPR131c
Length=251
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 56/98 (57%), Gaps = 9/98 (9%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQL------PTVAQDGQGKLVG 79
+ ++P + DLF+ ++N L EN+ ++++F + + WP L TV + + G
Sbjct 58 TTIQPFEPVDLFKTNNVNLDILTENFPLEFYFEYMIIWPDLFFKSSEMTVDPTFKHNISG 117
Query 80 YVLAKLEDENRQFQKPHGHVTSVAVMRQNRKLGLASKL 117
Y++AK E + ++ H H+T+V V + R++ LASKL
Sbjct 118 YMMAKTEGKTTEW---HTHITAVTVAPRFRRISLASKL 152
> SPCC16C4.12
Length=180
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 5/86 (5%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQD--GQGKLVGYVLAKLEDENRQF 92
DLF ++N L E + + ++ + WP L V + L+GY++ K E +++
Sbjct 11 DLFSFNNINLDPLTETFNISFYLSYLNKWPSLCVVQESDLSDPTLMGYIMGKSEGTGKEW 70
Query 93 QKPHGHVTSVAVMRQNRKLGLASKLM 118
H HVT++ V +R+LGLA +M
Sbjct 71 ---HTHVTAITVAPNSRRLGLARTMM 93
> 7298032_2
Length=203
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 49/100 (49%), Gaps = 7/100 (7%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLA-KLEDENRQFQ 93
DLF+ ++ L E Y + + L P+L A L+G++L ++ED F
Sbjct 10 DLFKFNNIVMDPLAEVYSLPFLLPKILEHPELVLAADAPDNSLMGFILGTRVEDATESFG 69
Query 94 KP------HGHVTSVAVMRQNRKLGLASKLMRLSQRAMQK 127
HGH++++AV + RKLGL ++L+ + M +
Sbjct 70 DAKTMTWNHGHISALAVAQDYRKLGLGTRLLTTVRDMMDR 109
> At2g38130
Length=190
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 45/82 (54%), Gaps = 4/82 (4%)
Query 46 NLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVM 105
L E Y + + + WPQL +A +GK VG ++ K+ D + F+ G++ + V+
Sbjct 37 ELSEPYSIFTYRYFVYLWPQLCFLAFH-KGKCVGTIVCKMGDHRQTFR---GYIAMLVVI 92
Query 106 RQNRKLGLASKLMRLSQRAMQK 127
+ R G+AS+L+ + +AM +
Sbjct 93 KPYRGRGIASELVTRAIKAMME 114
> ECU06g0460
Length=181
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGY 80
DL E+QHLN N EN+++ F +LS + D GK+VGY
Sbjct 33 DLLEVQHLNIRNSTENFLLGT-FLSTLSTSYGTSFVADLGGKIVGY 77
> CE07700
Length=278
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 6/77 (7%)
Query 51 YVMKYFFFHSLSWPQLPTVAQD-GQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVMRQNR 109
Y +YF + WP+ +A D +G VL KLE + + + G++ +AV R
Sbjct 124 YTYRYFLHN---WPEYCFLAYDQTNNTYIGAVLCKLELD--MYGRCKGYLAMLAVDESCR 178
Query 110 KLGLASKLMRLSQRAMQ 126
+LG+ ++L+R + AMQ
Sbjct 179 RLGIGTRLVRRALDAMQ 195
> SPBC15D4.06
Length=150
Score = 34.7 bits (78), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 6/76 (7%)
Query 51 YVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVMRQNRK 110
YV +YF WP+ VA D + +G V+ K +D +R G++ +A++++ R
Sbjct 30 YVYRYFVHQ---WPEFSFVALDND-RFIGAVICK-QDVHRG-TTLRGYIAMLAIVKEYRG 83
Query 111 LGLASKLMRLSQRAMQ 126
G+A+KL + S M+
Sbjct 84 QGIATKLTQASLDVMK 99
> YPR051w
Length=176
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 41/81 (50%), Gaps = 9/81 (11%)
Query 51 YVMKYFFFHSLSWPQLPTVAQDGQGKL----VGYVLAKLEDENRQFQKPHGHVTSVAVMR 106
YV +YF WP+L +A D + +G ++ K+ D +R + G++ +AV
Sbjct 34 YVYRYFLNQ---WPELTYIAVDNKSGTPNIPIGCIVCKM-DPHRNV-RLRGYIGMLAVES 88
Query 107 QNRKLGLASKLMRLSQRAMQK 127
R G+A KL+ ++ MQ+
Sbjct 89 TYRGHGIAKKLVEIAIDKMQR 109
> Hs22047261
Length=628
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 24 MCSLVRPSDVFDLFEMQHLNFVNL--PENYVMKYFFFHSLSWPQL 66
+C+L+RP F +FEM L+ + PEN +M ++ + L P L
Sbjct 40 VCALLRPHLSFPIFEMTKLDLLKCLSPENVMMLHYSWRMLKVPFL 84
> 7290141
Length=402
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 38/75 (50%), Gaps = 7/75 (9%)
Query 51 YVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVMRQNRK 110
Y +YF + +WP+L +A + VG ++ KL+ G++ +AV ++ RK
Sbjct 283 YTYRYFIY---NWPKLCFLASH-DNQYVGAIVCKLD---MHMNVRRGYIAMLAVRKEYRK 335
Query 111 LGLASKLMRLSQRAM 125
L + + L+ + AM
Sbjct 336 LKIGTTLVTKAIEAM 350
> CE13585
Length=891
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 83 AKLEDENR--QFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQR 123
AKLE++NR Q + PH + +++ + G+ S++ ++S R
Sbjct 133 AKLEEQNRKEQAKNPHASAETEPAVKKKKIEGVGSRIPKISDR 175
> CE18636_2
Length=2051
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 45 VNLPENYVMKYFFFHSLSWPQLPTV 69
++LP YV Y+FFH +PQL V
Sbjct 1001 IHLPAAYVRDYYFFHRSFYPQLTLV 1025
> Hs20541823
Length=198
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Query 15 AAFRRLPAKMCSLVRPSDVFDLF-EMQHLNFVNLPENYVMKYFF---FHSLSWPQLPTVA 70
A+R A C+L D F F + Q ++F N+ E+ ++KY++ + PQ P
Sbjct 36 TAYRSEQALSCTLEH--DTFKAFYQHQVIDFHNVVESLIIKYYYPDLTGLIHLPQAPAAK 93
Query 71 QDGQGK 76
+GK
Sbjct 94 SKARGK 99
> 7290142
Length=383
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 39/76 (51%), Gaps = 9/76 (11%)
Query 51 YVMKYFFFHSLSWPQLPTVA-QDGQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVMRQNR 109
Y +YF + +WP+L +A D Q VG ++ KL + G++ +AV ++ R
Sbjct 264 YTYRYFIY---NWPKLCFLASHDNQ--YVGAIVCKL---DMHMNVRRGYIAMLAVRKEYR 315
Query 110 KLGLASKLMRLSQRAM 125
KL + + L+ + AM
Sbjct 316 KLKIGTTLVTKAIEAM 331
> At1g55150
Length=501
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 6/61 (9%)
Query 59 HSLSWP------QLPTVAQDGQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVMRQNRKLG 112
S WP L +A+ G GK + Y+L + N Q HG V V+ R+L
Sbjct 126 QSQGWPMAMKGRDLIGIAETGSGKTLSYLLPAIVHVNAQPMLAHGDGPIVLVLAPTRELA 185
Query 113 L 113
+
Sbjct 186 V 186
> At1g35580
Length=544
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 41 HLNFVNLPENYVMKYFFFHSLSWPQL 66
H+ ++L E+ +MK S SWPQL
Sbjct 519 HIGMISLEEDKLMKPVIKRSASWPQL 544
> At3g09570
Length=439
Score = 26.9 bits (58), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 40 QHLNFVNLPENYVMKYFFFHSLSWP 64
Q+ NF L NYV+ F FH LS P
Sbjct 93 QNPNFCVLDSNYVLHLFTFHDLSPP 117
Lambda K H
0.329 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40