bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2063_orf1
Length=160
Score E
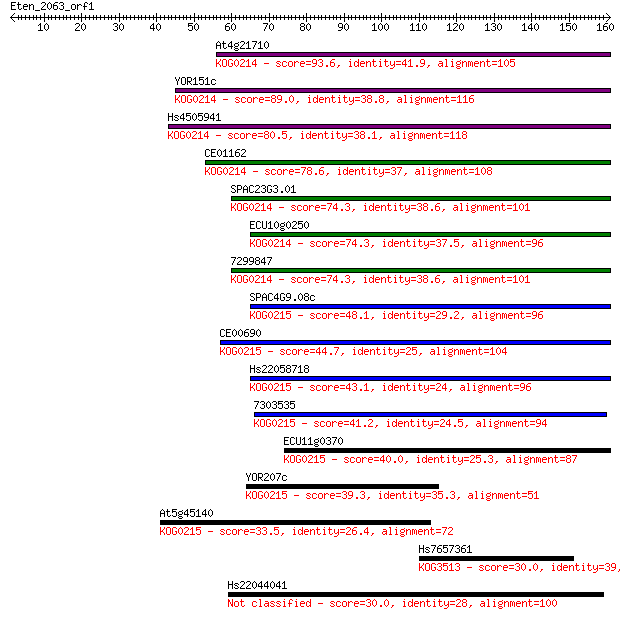
Sequences producing significant alignments: (Bits) Value
At4g21710 93.6 1e-19
YOR151c 89.0 4e-18
Hs4505941 80.5 1e-15
CE01162 78.6 4e-15
SPAC23G3.01 74.3 8e-14
ECU10g0250 74.3 9e-14
7299847 74.3 9e-14
SPAC4G9.08c 48.1 7e-06
CE00690 44.7 7e-05
Hs22058718 43.1 2e-04
7303535 41.2 0.001
ECU11g0370 40.0 0.002
YOR207c 39.3 0.004
At5g45140 33.5 0.17
Hs7657361 30.0 1.7
Hs22044041 30.0 2.0
> At4g21710
Length=1188
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 67/106 (63%), Gaps = 1/106 (0%)
Query 56 DEQQQEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYR 115
D+ +EI ED WAV+ ++FE KGLV QQ++SF++F +QEI+D IEIRP Q+
Sbjct 15 DDDDEEITQEDAWAVISAYFEEKGLVRQQLDSFDEFIQNTMQEIVDESADIEIRPESQHN 74
Query 116 PEEDVEL-DVRYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
P + + Y++ FGQ+ L++P E +G L+P+ +R RNL
Sbjct 75 PGHQSDFAETIYKISFGQIYLSKPMMTESDGETATLFPKAARLRNL 120
> YOR151c
Length=1224
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 45/118 (38%), Positives = 70/118 (59%), Gaps = 3/118 (2%)
Query 45 NDELQQQEDQH--DEQQQEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDA 102
N E ED + +++ I +ED+WAV+ +FF KGLVSQQ++SFN F Y LQ+II
Sbjct 6 NSEKYYDEDPYGFEDESAPITAEDSWAVISAFFREKGLVSQQLDSFNQFVDYTLQDIICE 65
Query 103 HPPIEIRPSPQYRPEEDVELDVRYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
+ + Q+ E D + +Y + FG++ + +P E +G+ L+PQE+R RNL
Sbjct 66 DSTLILEQLAQHTTESD-NISRKYEISFGKIYVTKPMVNESDGVTHALYPQEARLRNL 122
> Hs4505941
Length=1174
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 45/118 (38%), Positives = 72/118 (61%), Gaps = 2/118 (1%)
Query 43 DENDELQQQEDQHDEQQQEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDA 102
D ++++Q ED DE ++ E W V+ S+F+ KGLV QQ++SF++F +Q I++
Sbjct 3 DADEDMQYDEDD-DEITPDLWQEACWIVISSYFDEKGLVRQQLDSFDEFIQMSVQRIVED 61
Query 103 HPPIEIRPSPQYRPEEDVELDVRYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
PPI+++ Q+ E VE RY LKF Q+ L++P E++G + P E+R RNL
Sbjct 62 APPIDLQAEAQHASGE-VEEPPRYLLKFEQIYLSKPTHWERDGAPSPMMPNEARLRNL 118
> CE01162
Length=1193
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 64/108 (59%), Gaps = 1/108 (0%)
Query 53 DQHDEQQQEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSP 112
D DE E E W V+ ++F+ KGLV QQ++SF++F +Q I++ PP+E++
Sbjct 19 DDSDEISAEAWQEACWVVISAYFDEKGLVRQQLDSFDEFVQMNVQRIVEDSPPVELQSEN 78
Query 113 QYRPEEDVELDVRYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
Q+ D+E ++ LKF Q+ L++P EK+G + P E+R RNL
Sbjct 79 QHL-GTDMENPAKFSLKFNQIYLSKPTHWEKDGAPMPMMPNEARLRNL 125
> SPAC23G3.01
Length=728
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 57/103 (55%), Gaps = 5/103 (4%)
Query 60 QEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRPEED 119
+ + ED W V+ SFFE L QQ+ SF++F +QEI+D + + QY
Sbjct 10 ETLTQEDCWTVISSFFEETSLARQQLFSFDEFVQNTMQEIVDDDSTLTL---DQYAQHTG 66
Query 120 VELDV--RYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
+ DV RY + FGQ+ L+RP E +G ++PQE+R RNL
Sbjct 67 AQGDVTRRYEINFGQIYLSRPTMTEADGSTTTMFPQEARLRNL 109
> ECU10g0250
Length=1141
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 36/97 (37%), Positives = 59/97 (60%), Gaps = 5/97 (5%)
Query 65 EDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRPEEDVELDV 124
E +W+++ SFFE KGLV QQ++SF+ F K+QE+I+ P I + Q P +E
Sbjct 8 EHSWSIISSFFEQKGLVRQQLDSFDQFVKTKMQEVINESPVIIV----QSAPTAGIETQK 63
Query 125 RYRLKFGQLSLNRPAA-QEKEGMQKNLWPQESRTRNL 160
R ++FGQ+ +++P E +G ++P E+R R+L
Sbjct 64 RIAVRFGQIYVSKPPVYTESDGRTITVFPNEARIRDL 100
> 7299847
Length=1176
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 39/101 (38%), Positives = 61/101 (60%), Gaps = 1/101 (0%)
Query 60 QEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRPEED 119
E+ E W V+ ++F+ KGLV QQ++SF++F +Q I++ P IE++ Q+ E
Sbjct 20 HELWQEACWIVINAYFDEKGLVRQQLDSFDEFIQMSVQRIVEDSPAIELQAEAQHTSGE- 78
Query 120 VELDVRYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
VE R+ LKF Q+ L++P EK+G + P E+R RNL
Sbjct 79 VETPPRFSLKFEQIYLSKPTHWEKDGSPSPMMPNEARLRNL 119
> SPAC4G9.08c
Length=1165
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 28/98 (28%), Positives = 49/98 (50%), Gaps = 16/98 (16%)
Query 65 EDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRPEEDVELDV 124
ED W ++ +F + KGLV Q ++S+N F L++I+ A+ E V DV
Sbjct 48 EDKWQLLPAFLKVKGLVKQHLDSYNYFVDVDLKKIVQAN--------------EKVTSDV 93
Query 125 R--YRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
+ LK+ + + P + + +Q ++ P E R R+L
Sbjct 94 EPWFYLKYLDIRVGAPVRTDADAIQASISPHECRLRDL 131
> CE00690
Length=1207
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 26/105 (24%), Positives = 54/105 (51%), Gaps = 13/105 (12%)
Query 57 EQQQEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRP 116
+ Q+ + ED W +V +F + +GLV Q + SF+ F +++ I+ ++ I +P
Sbjct 71 KDQKCMPLEDKWLLVPAFLKVRGLVKQHLVSFDHFVQEEIRSIMLSNQKITSDANPN--- 127
Query 117 EEDVELDVRYRLKFGQLSLNRPAAQEKEGMQKN-LWPQESRTRNL 160
+ LK+ + + +P+++E M + + PQE R R++
Sbjct 128 ---------FYLKYLDIRIGKPSSEEGLNMTHDKITPQECRLRDM 163
> Hs22058718
Length=273
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 48/96 (50%), Gaps = 12/96 (12%)
Query 65 EDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRPEEDVELDV 124
E+ W ++ +F + KGLV Q ++SFN F + ++++I+ A+ + P
Sbjct 24 EEKWRLLPAFLKVKGLVKQHIDSFNYFINVEIKKIMKANEKVTSDADPM----------- 72
Query 125 RYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRNL 160
+ LK+ + + P +E + + + P E R R++
Sbjct 73 -WYLKYLNIYVGLPDVEESFNVTRPVSPHECRLRDM 107
> 7303535
Length=1137
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 23/94 (24%), Positives = 44/94 (46%), Gaps = 12/94 (12%)
Query 66 DTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRPEEDVELDVR 125
+ W +V +F + KGLV Q ++SFN F + +++I+ A+ + P
Sbjct 33 EKWKLVPAFLQVKGLVKQHIDSFNHFINVDIKKIVKANELVTSGADPL------------ 80
Query 126 YRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTRN 159
+ LK+ + + +P + + K P E R R+
Sbjct 81 FYLKYLDVRVGKPDIDDGFNITKATTPHECRLRD 114
> ECU11g0370
Length=1110
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 46/87 (52%), Gaps = 13/87 (14%)
Query 74 FFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQYRPEEDVELDVRYRLKFGQL 133
FF KGLV Q +ES++ F ++++ ++ A+ + D ++D + LK+ +
Sbjct 13 FFNEKGLVRQHIESYDYFVDHEIKALVRANQIV------------DSDIDHTFYLKYLDI 60
Query 134 SLNRPAAQEKEGMQKNLWPQESRTRNL 160
+ P+ +E + N++P E R R++
Sbjct 61 RVAMPSVEENM-VNYNVFPIECRLRDI 86
> YOR207c
Length=1149
Score = 39.3 bits (90), Expect = 0.004, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 64 SEDTWAVVGSFFEGKGLVSQQVESFNDFASYKLQEIIDAHPPIEIRPSPQY 114
++D W ++ +F + KGLV Q ++SFN F L++II A+ I P++
Sbjct 41 AQDKWHLLPAFLKVKGLVKQHLDSFNYFVDTDLKKIIKANQLILSDVDPEF 91
> At5g45140
Length=1194
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 19/77 (24%), Positives = 39/77 (50%), Gaps = 5/77 (6%)
Query 41 VGDENDELQQQEDQHDEQQQEID-----SEDTWAVVGSFFEGKGLVSQQVESFNDFASYK 95
+G + ++L D H ++++ + D + +V F + +GLV Q ++SFN F +
Sbjct 22 MGLDQEDLDLTNDDHFIDKEKLSAPIKSTADKFQLVPEFLKVRGLVKQHLDSFNYFINVG 81
Query 96 LQEIIDAHPPIEIRPSP 112
+ +I+ A+ I P
Sbjct 82 IHKIVKANSRITSTVDP 98
> Hs7657361
Length=1028
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 5/46 (10%)
Query 110 PSPQYR-----PEEDVELDVRYRLKFGQLSLNRPAAQEKEGMQKNL 150
PSP YR + D + YRL G L++N P + GM + L
Sbjct 56 PSPHYRWKQNGTDIDFTMSYHYRLDGGSLAINSPHTDQDIGMYQCL 101
> Hs22044041
Length=2375
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 3/103 (2%)
Query 59 QQEIDSEDTWAVVGSFFEGKGLVSQQVESFNDFA---SYKLQEIIDAHPPIEIRPSPQYR 115
+++I E A+V + +E KGL V +N F +YK +II + P + R
Sbjct 999 KEKIPEELGLALVLTIYEAKGLEFDDVLLYNFFTDSEAYKEWKIISSFTPTSTDSREENR 1058
Query 116 PEEDVELDVRYRLKFGQLSLNRPAAQEKEGMQKNLWPQESRTR 158
P +V LD + L +N + G K L+ +R R
Sbjct 1059 PLVEVPLDKPGSSQGRSLMVNPEMYKLLNGELKQLYTAITRAR 1101
Lambda K H
0.312 0.132 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2172509160
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40