bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2075_orf3
Length=226
Score E
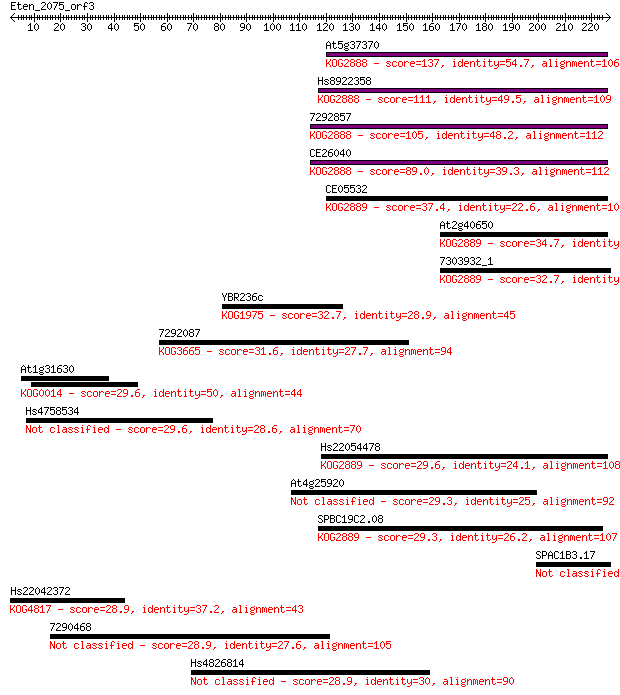
Sequences producing significant alignments: (Bits) Value
At5g37370 137 1e-32
Hs8922358 111 1e-24
7292857 105 6e-23
CE26040 89.0 8e-18
CE05532 37.4 0.023
At2g40650 34.7 0.18
7303932_1 32.7 0.58
YBR236c 32.7 0.62
7292087 31.6 1.4
At1g31630 29.6 5.1
Hs4758534 29.6 5.5
Hs22054478 29.6 5.7
At4g25920 29.3 5.9
SPBC19C2.08 29.3 7.6
SPAC1B3.17 28.9 7.7
Hs22042372 28.9 7.9
7290468 28.9 8.1
Hs4826814 28.9 9.7
> At5g37370
Length=393
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 58/106 (54%), Positives = 76/106 (71%), Gaps = 0/106 (0%)
Query 120 VNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYKLF 179
+ ++L NI+SS+YFK L+ K+++EV+DE+ +H EP+ G+ R PST +C LYK F
Sbjct 15 LEKVLSMNILSSDYFKELYGLKTYHEVIDEIYNQVNHVEPWMGGNCRGPSTAYCLLYKFF 74
Query 180 TLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
T+KLT KQMH LL H +SPY+R GFLYLRYV A LW WYEPY
Sbjct 75 TMKLTVKQMHGLLKHTDSPYIRAVGFLYLRYVADAKTLWTWYEPYI 120
> Hs8922358
Length=546
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 54/128 (42%), Positives = 74/128 (57%), Gaps = 19/128 (14%)
Query 117 TYNVNQLLRGNIMSSEYFK-SLHQFKSFNEVVDELAAFADHAEPYCSGSTRA-------- 167
T N+N ++ NI+SS YFK L++ K+++EVVDE+ H EP+ GS +
Sbjct 55 TMNLNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTGMCG 114
Query 168 ----------PSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQL 217
ST FC LYKLFTLKLT KQ+ L+ H +SPY+R GF+Y+RY P L
Sbjct 115 GVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPPTDL 174
Query 218 WKWYEPYF 225
W W+E +
Sbjct 175 WDWFESFL 182
> 7292857
Length=398
Score = 105 bits (262), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 75/131 (57%), Gaps = 19/131 (14%)
Query 114 DSTTYNVNQLLRGNIMSSEYFK-SLHQFKSFNEVVDELAAFADHAEPYCSGSTRAP---- 168
+ ++ N+N L+ NI SS YFK L + K+++EVVDE+ H EP+ GS +
Sbjct 29 NESSMNLNALILANIQSSSYFKVHLFKLKTYHEVVDEIYYQVKHMEPWERGSRKTSGQTG 88
Query 169 --------------STLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPA 214
ST +C LYKL+TL+LT KQ++ LLNH +S Y+R GF+YLRY P
Sbjct 89 MCGGVRGVGAGGIVSTAYCLLYKLYTLRLTRKQINGLLNHTDSAYIRALGFMYLRYTQPP 148
Query 215 DQLWKWYEPYF 225
L+ WYE Y
Sbjct 149 GDLYDWYEDYL 159
> CE26040
Length=327
Score = 89.0 bits (219), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 69/131 (52%), Gaps = 19/131 (14%)
Query 114 DSTTYNVNQLLRGNIMSSEYFKS-LHQFKSFNEVVDELAAFADHAEPYCSGSTR------ 166
+ T N+N L+ NI S Y+K+ L + +F +++++ H EP+ G+ R
Sbjct 60 NKVTMNLNTLVLENIRESYYYKNNLVEIDNFQTLIEQIFYQVKHLEPWEKGTRRLQGMTG 119
Query 167 ------------APSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPA 214
S+ +C LY+LF LK++ KQ+ +LN R+S Y+R GF+Y+RY P
Sbjct 120 MCGGVRGVGAGGVVSSAYCLLYRLFNLKISRKQLISMLNSRQSVYIRGLGFMYIRYTQPP 179
Query 215 DQLWKWYEPYF 225
LW W EPY
Sbjct 180 ADLWYWLEPYL 190
> CE05532
Length=320
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 24/106 (22%), Positives = 49/106 (46%), Gaps = 2/106 (1%)
Query 120 VNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYKLF 179
V +++R I S Y+K H F E+V + + +G+ + P+ C K+
Sbjct 21 VEKIIRQRIYDSMYWKE-HCFALTAELVVDKGMDLRYIGGIYAGNIK-PTPFLCLALKML 78
Query 180 TLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
++ + + + E Y+R G +YLR + +++K+ EP +
Sbjct 79 QIQPDKDIVLEFIQQEEFKYIRALGAMYLRLTFDSTEIYKYLEPLY 124
> At2g40650
Length=363
Score = 34.7 bits (78), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 14/63 (22%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
G +R P+ C + K+ ++ ++ + + + + YVR G YLR ++++ E
Sbjct 62 GGSRKPTPFLCLILKMLQIQPEKEIVVEFIKNDDYKYVRILGAFYLRLTGTDVDVYRYLE 121
Query 223 PYF 225
P +
Sbjct 122 PLY 124
> 7303932_1
Length=355
Score = 32.7 bits (73), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 16/64 (25%), Positives = 28/64 (43%), Gaps = 0/64 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
G P+ C K+ ++ + + + + E YVR G YLR A +K+ E
Sbjct 62 GGNIKPTQFLCLTLKMLQIQPEKDIVVEFIKNEEFKYVRALGAFYLRLTGAALDCYKYLE 121
Query 223 PYFL 226
P ++
Sbjct 122 PLYI 125
> YBR236c
Length=436
Score = 32.7 bits (73), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 81 RKYKSFTQQMGVLQQRAIAAEEETQRKNMVQMTDSTTYNVNQLLR 125
R+++ + Q+ + +QRA EE +++ ++MT + + NV+Q++R
Sbjct 79 RRHERYDQEERLRKQRAQKLREEQLKRHEIEMTANRSINVDQIVR 123
> 7292087
Length=758
Score = 31.6 bits (70), Expect = 1.4, Method: Composition-based stats.
Identities = 26/106 (24%), Positives = 45/106 (42%), Gaps = 14/106 (13%)
Query 57 KGDEEEAIARCHLHTKPQLSCKFCRKYKSFTQQMGVLQQRAIAAEEETQR---------- 106
K ++E + R ++ L+C+ + K F ++GV+ ++ R
Sbjct 481 KTEQEGFVQRIAIYLLNTLACQVDGRQKLFLGELGVVSTMFTLIKDRLTRSVFDDVMEVA 540
Query 107 -KNMVQMTDSTTYNVNQLLRGNIMSSEYF-KSLHQFKSFNEVVDEL 150
M +TD T N + L G M EYF K LH F +E++ +
Sbjct 541 WSTMWNVTDETAINCKRFLDGRGM--EYFLKCLHTFPDRDELLRNM 584
> At1g31630
Length=339
Score = 29.6 bits (65), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 5 AGVAPLFYGATSAATAAVAAAAVAPIAAAAAAP 37
AG APL A+ AVA AP+A A A P
Sbjct 189 AGAAPLAVAGAGASPLAVAGVGAAPLAVAGAGP 221
Score = 29.3 bits (64), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 20/40 (50%), Gaps = 3/40 (7%)
Query 9 PLFYGATSAATAAVAAAAVAPIAAAAAAPVAVAPLAAAAA 48
PL AA AVA A +P+A A V APLA A A
Sbjct 183 PLVVAGAGAAPLAVAGAGASPLAVAG---VGAAPLAVAGA 219
> Hs4758534
Length=473
Score = 29.6 bits (65), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 33/70 (47%), Gaps = 2/70 (2%)
Query 7 VAPLFYGATSAATAAVAAAAVAPIAAAAAAPVAVAPLAAAAADVGAADKDKGDEEEAIAR 66
++P GA A AA + P AAA ++P+A A +++G + G + + R
Sbjct 108 LSPSGMGAMGAQQAASMMNGLGPYAAAMNP--CMSPMAYAPSNLGRSRAGGGGDAKTFKR 165
Query 67 CHLHTKPQLS 76
+ H KP S
Sbjct 166 SYPHAKPPYS 175
> Hs22054478
Length=264
Score = 29.6 bits (65), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 46/109 (42%), Gaps = 4/109 (3%)
Query 118 YNVNQLLRGNIMSSEYFKSLHQFKSFNE-VVDELAAFADHAEPYCSGSTRAPSTLFCCLY 176
Y V +++R I S+++K F E VVD Y G P+ C +
Sbjct 19 YLVGKIIRMRICESKHWKE-ECFGLMAELVVDNAMELMFVGGEY--GGNIKPTPFLCLIL 75
Query 177 KLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
K+ ++ + + + + YV G LY+R + A +K+ EP +
Sbjct 76 KMLQIQSEKGITAEFIENEDFKYVHMLGALYMRLMGTAIDCYKYLEPLY 124
> At4g25920
Length=365
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 42/100 (42%), Gaps = 12/100 (12%)
Query 107 KNMVQMTDSTTYNVNQLL----RGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCS 162
+++ + ++T YNV + G S Y K + + E + +L F H+E +C
Sbjct 249 EDLEDIDNTTLYNVTKKFMVFKEGEKPSCVYTKKM----IYTEDIGDLCIFVGHSEAFCV 304
Query 163 GSTRAPSTLFCCL----YKLFTLKLTEKQMHMLLNHRESP 198
++ +P C+ Y LT K M L ++P
Sbjct 305 PASSSPGLKPNCIYFVGYNFGVYDLTTKTCTMFLTKDDNP 344
> SPBC19C2.08
Length=210
Score = 29.3 bits (64), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 28/109 (25%), Positives = 47/109 (43%), Gaps = 7/109 (6%)
Query 117 TYNVNQLLRGNIMSSEYFKSLHQFKSFN--EVVDELAAFADHAEPYCSGSTRAPSTLFCC 174
T+ + ++LR I+ S Y+K Q N +VD Y G+ R P+ C
Sbjct 22 TFLIGKILRERIVDSIYWK--EQCFGLNACSLVDRAVRLEYIGGQY--GNQR-PTEFICL 76
Query 175 LYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEP 223
LYKL + ++ + L+ E Y+R Y+R ++ + EP
Sbjct 77 LYKLLQIAPEKEIIQQYLSIPEFKYLRALAAFYVRLTWDDVEVHQTLEP 125
> SPAC1B3.17
Length=537
Score = 28.9 bits (63), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 1/29 (3%)
Query 199 YVRCTGFLYLRYVHP-ADQLWKWYEPYFL 226
Y RC + YV P AD L +WYEP+F+
Sbjct 462 YFRCLHDKSVEYVCPFADVLGRWYEPWFV 490
> Hs22042372
Length=2698
Score = 28.9 bits (63), Expect = 7.9, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 1 AAPHAGVAPLFYGAT---SAATAAVAAAAVAPIAAAAAAPVAVAPL 43
+ P + APL T ++ +A V A+ +AP+ A+ +APV +PL
Sbjct 1758 SVPASTSAPLPATLTPVPASTSAPVPASTLAPVLASTSAPVPASPL 1803
> 7290468
Length=188
Score = 28.9 bits (63), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 9/110 (8%)
Query 16 SAATAAVAAAAVAPIAAAAAAPVAVAPLAAA-AADVGAADKDKGDEEEAIARCHLHT--K 72
A AAV A V+ + A +A L + +D+ A D D ++ + HL+T K
Sbjct 75 GALQAAVDAHRVSDVNQVDQAAKDIASLRSKLGSDLRPAILDSNDVKQCLE--HLNTTHK 132
Query 73 PQLSCKFCRKYKSFTQQMGVLQ--QRAIAAEEETQRKNMVQMTDSTTYNV 120
P+L+ CR+ + F Q L+ + A+ E M+Q D ++
Sbjct 133 PRLN--LCRQQREFAQNQEALRSLRTAVDGLENGMEMGMIQAMDRLVEDL 180
> Hs4826814
Length=1300
Score = 28.9 bits (63), Expect = 9.7, Method: Composition-based stats.
Identities = 27/93 (29%), Positives = 47/93 (50%), Gaps = 13/93 (13%)
Query 69 LHTKPQLSCKFCRKYKS-FTQQMGVLQ--QRAIAAEEETQRKNMVQMTDSTTYNVNQLLR 125
+HT QL C+ KYKS + G+LQ QR++ EE + + D + + Q ++
Sbjct 1099 MHTLLQLECE---KYKSVLAETEGILQKLQRSVEQEENKWKVKV----DESHKTIKQ-MQ 1150
Query 126 GNIMSSEYFKSLHQFKSFNEVVDELAAFADHAE 158
+ SSE + L + +S N+ ++ L +H E
Sbjct 1151 SSFTSSE--QELERLRSENKDIENLRREREHLE 1181
Lambda K H
0.321 0.129 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4312910206
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40