bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2081_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
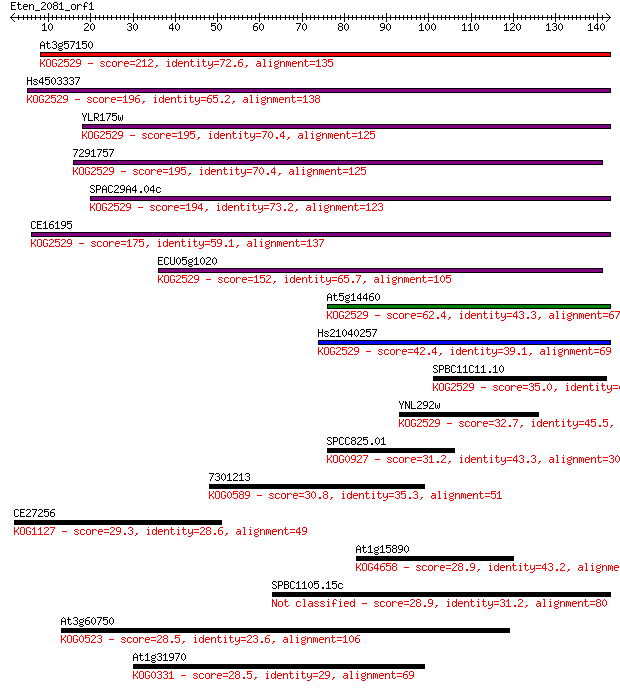
At3g57150 212 2e-55
Hs4503337 196 9e-51
YLR175w 195 2e-50
7291757 195 2e-50
SPAC29A4.04c 194 4e-50
CE16195 175 3e-44
ECU05g1020 152 2e-37
At5g14460 62.4 3e-10
Hs21040257 42.4 3e-04
SPBC11C11.10 35.0 0.042
YNL292w 32.7 0.21
SPCC825.01 31.2 0.60
7301213 30.8 0.90
CE27256 29.3 2.4
At1g15890 28.9 3.3
SPBC1105.15c 28.9 3.3
At3g60750 28.5 3.9
At1g31970 28.5 3.9
> At3g57150
Length=565
Score = 212 bits (539), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 98/135 (72%), Positives = 114/135 (84%), Gaps = 0/135 (0%)
Query 8 EKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPL 67
+K + AA ++I+P S P IDTS+WP+LLKNYDRLNVRTGHYTP+ G SPL RPL
Sbjct 15 DKTENDAADTGDYMIKPQSFTPAIDTSQWPILLKNYDRLNVRTGHYTPISAGHSPLKRPL 74
Query 68 QQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLV 127
Q+++ YG + LDKPANPSSHEVVAW+K+ILRVEKTGHSGTLDPKVTG L+VCI+RATRLV
Sbjct 75 QEYIRYGVINLDKPANPSSHEVVAWIKRILRVEKTGHSGTLDPKVTGNLIVCIDRATRLV 134
Query 128 KSQQSAGKEYVAVAR 142
KSQQ AGKEYV VAR
Sbjct 135 KSQQGAGKEYVCVAR 149
> Hs4503337
Length=514
Score = 196 bits (499), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 90/138 (65%), Positives = 112/138 (81%), Gaps = 0/138 (0%)
Query 5 TMGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLA 64
++ E++ + AE F+I+P S +DTS+WPLLLKN+D+LNVRT HYTPL GS+PL
Sbjct 21 SLPEEDVAEIQHAEEFLIKPESKVAKLDTSQWPLLLKNFDKLNVRTTHYTPLACGSNPLK 80
Query 65 RPLQQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRAT 124
R + ++ G + LDKP+NPSSHEVVAW+++ILRVEKTGHSGTLDPKVTGCL+VCI RAT
Sbjct 81 REIGDYIRTGFINLDKPSNPSSHEVVAWIRRILRVEKTGHSGTLDPKVTGCLIVCIERAT 140
Query 125 RLVKSQQSAGKEYVAVAR 142
RLVKSQQSAGKEYV + R
Sbjct 141 RLVKSQQSAGKEYVGIVR 158
> YLR175w
Length=483
Score = 195 bits (496), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 88/125 (70%), Positives = 106/125 (84%), Gaps = 0/125 (0%)
Query 18 EGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGSLL 77
E FVI+P + DTS+WPLLLKN+D+L VR+GHYTP+P GSSPL R L+ +++ G +
Sbjct 4 EDFVIKPEAAGASTDTSEWPLLLKNFDKLLVRSGHYTPIPAGSSPLKRDLKSYISSGVIN 63
Query 78 LDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGKEY 137
LDKP+NPSSHEVVAW+K+ILR EKTGHSGTLDPKVTGCL+VCI+RATRLVKSQQ AGKEY
Sbjct 64 LDKPSNPSSHEVVAWIKRILRCEKTGHSGTLDPKVTGCLIVCIDRATRLVKSQQGAGKEY 123
Query 138 VAVAR 142
V + R
Sbjct 124 VCIVR 128
> 7291757
Length=508
Score = 195 bits (495), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 88/125 (70%), Positives = 108/125 (86%), Gaps = 0/125 (0%)
Query 16 KAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGS 75
K F I+P+S +DTS+WPLLLKN+D+LN+R+ HYTPL GSSPL R +++++ G
Sbjct 30 KQGNFQIKPSSKIAELDTSQWPLLLKNFDKLNIRSNHYTPLAHGSSPLNRDIKEYMKTGF 89
Query 76 LLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGK 135
+ LDKP+NPSSHEVVAW+KKIL+VEKTGHSGTLDPKVTGCL+VCI+RATRLVKSQQSAGK
Sbjct 90 INLDKPSNPSSHEVVAWIKKILKVEKTGHSGTLDPKVTGCLIVCIDRATRLVKSQQSAGK 149
Query 136 EYVAV 140
EYVA+
Sbjct 150 EYVAI 154
> SPAC29A4.04c
Length=474
Score = 194 bits (494), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 90/124 (72%), Positives = 110/124 (88%), Gaps = 1/124 (0%)
Query 20 FVIQPTST-APPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGSLLL 78
F+I+P +T A IDT++WPLLLKN+D+L VRTGHYTP+P G++PL RP+ ++++ G + L
Sbjct 10 FMIKPEATSASKIDTAEWPLLLKNFDKLLVRTGHYTPIPCGNNPLKRPIAEYVSSGVINL 69
Query 79 DKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGKEYV 138
DKPANPSSHEVVAW+KKILRVEKTGHSGTLDPKVTGCL++C +RATRLVKSQQSAGKEYV
Sbjct 70 DKPANPSSHEVVAWVKKILRVEKTGHSGTLDPKVTGCLIICNDRATRLVKSQQSAGKEYV 129
Query 139 AVAR 142
V R
Sbjct 130 CVLR 133
> CE16195
Length=445
Score = 175 bits (443), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 81/146 (55%), Positives = 107/146 (73%), Gaps = 9/146 (6%)
Query 6 MGEKEKK---------KAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPL 56
MG+K+K+ +A + F + ++ +D S+WPLLLKNYD+LNVRT HYTP
Sbjct 1 MGKKDKRSKLEGDDLAEAQQKGSFQLPSSNETAKLDASQWPLLLKNYDKLNVRTNHYTPH 60
Query 57 PFGSSPLARPLQQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCL 116
G SPL R ++ +++ G LDKP+NPSSHEVV+W+K+ILR EKTGHSGTLDPKV+GCL
Sbjct 61 VEGVSPLKRDIKNYISSGFFNLDKPSNPSSHEVVSWIKRILRCEKTGHSGTLDPKVSGCL 120
Query 117 LVCINRATRLVKSQQSAGKEYVAVAR 142
+VCI+R TRL KSQQ AGKEY+ + +
Sbjct 121 IVCIDRTTRLAKSQQGAGKEYICIFK 146
> ECU05g1020
Length=357
Score = 152 bits (383), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 69/105 (65%), Positives = 84/105 (80%), Gaps = 0/105 (0%)
Query 36 WPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGSLLLDKPANPSSHEVVAWLKK 95
W LLL N DR+ VRTGHYTPL GS PL R +++++ YG + +DK NPSSHEVV W+K+
Sbjct 20 WDLLLANMDRMVVRTGHYTPLGCGSMPLRRRIEEYIKYGIINVDKSPNPSSHEVVTWVKE 79
Query 96 ILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGKEYVAV 140
IL+ EKTGHSGTLDP+V+G L +CI+RATRL KSQQS GKEYV V
Sbjct 80 ILKCEKTGHSGTLDPQVSGVLTICIDRATRLTKSQQSLGKEYVCV 124
> At5g14460
Length=540
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 44/67 (65%), Gaps = 0/67 (0%)
Query 76 LLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGK 135
LL++KP +S V L+++++V+K GH+GTLDP TG L+VC+ +AT++V Q K
Sbjct 324 LLVNKPKGWTSFTVCGKLRRLVKVKKVGHAGTLDPMATGLLIVCVGKATKVVDRYQGMIK 383
Query 136 EYVAVAR 142
Y V R
Sbjct 384 GYSGVFR 390
> Hs21040257
Length=349
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 37/85 (43%), Gaps = 16/85 (18%)
Query 74 GSLLLDKPANPSSHEVVAWLKKILRVE----------------KTGHSGTLDPKVTGCLL 117
G + KP P+S E++ LK+ L E K GH GTLD G L+
Sbjct 70 GVFAVHKPKGPTSAELLNRLKEKLLAEAGMPSPEWTKRKKQTLKIGHGGTLDSAARGVLV 129
Query 118 VCINRATRLVKSQQSAGKEYVAVAR 142
V I T+++ S S K Y A+
Sbjct 130 VGIGSGTKMLTSMLSGSKRYTAIGE 154
> SPBC11C11.10
Length=407
Score = 35.0 bits (79), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 101 KTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGKEYVAVA 141
K GH GTLDP +G L+V + T+ + S S K Y A A
Sbjct 107 KIGHGGTLDPLASGVLVVGLGTGTKQLSSLLSCMKTYRATA 147
> YNL292w
Length=403
Score = 32.7 bits (73), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Query 93 LKKILRVEKTGHSGTLDPKVTGCLLVCINRATR 125
L+K+ +V K GH GTLDP +G L++ I T+
Sbjct 60 LRKVSKV-KMGHGGTLDPLASGVLVIGIGAGTK 91
> SPCC825.01
Length=822
Score = 31.2 bits (69), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 76 LLLDKPANPSSHEVVAWLKKILRVEKTGHS 105
++LD+P N E VAWL++ L E GH+
Sbjct 442 MMLDEPTNHLDLEAVAWLEEYLTHEMEGHT 471
> 7301213
Length=841
Score = 30.8 bits (68), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query 48 VRTGHYTPLPFGSSPLARPLQQHLNYGSLLLDKPANPSSHEV-VAWLKKILR 98
+ G+YTP+P G + R L +L L ++ P P++ EV V W+ I R
Sbjct 316 IMAGNYTPVPSGYTSGLRSLMSNL----LQVEAPRRPTASEVLVYWIPLIFR 363
> CE27256
Length=959
Score = 29.3 bits (64), Expect = 2.4, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 8/49 (16%)
Query 2 YVDTMGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRT 50
++ GE++K++AA+A + ++ WP +L+N+D LN T
Sbjct 98 FMQNQGEEKKEQAAQARRRI--------SVELKLWPEILENFDVLNFNT 138
> At1g15890
Length=851
Score = 28.9 bits (63), Expect = 3.3, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 83 NPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVC 119
N SSHE + +KIL V K + D KV C L C
Sbjct 376 NSSSHEFPSMEEKILPVLKFSYDDLKDEKVKLCFLYC 412
> SPBC1105.15c
Length=300
Score = 28.9 bits (63), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 39/96 (40%), Gaps = 17/96 (17%)
Query 63 LARPLQQHLN---YGSLLLDKPANPSSHEVVAWLKKILRVEKTGH-------------SG 106
LA P + +LN Y SL KP NP S + WL +LR + H
Sbjct 52 LASP-ESNLNSDGYESLYSPKPNNPFSFKRRIWLHGVLRFHRPLHLFQTSECHELIQEES 110
Query 107 TLDPKVTGCLLVCINRATRLVKSQQSAGKEYVAVAR 142
P +T + C+ +++ L K ++V + R
Sbjct 111 VKSPSITKEVEACLPKSSFLTKPTSEKQTKHVYIRR 146
> At3g60750
Length=754
Score = 28.5 bits (62), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 42/106 (39%), Gaps = 22/106 (20%)
Query 13 KAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLN 72
+AA E ++PT+ + +D S + D + + LP G +P+A H+
Sbjct 65 RAAAVE--TVEPTTDSSIVDKSVNSIRFLAIDAVEKAKSGHPGLPMGCAPMA-----HIL 117
Query 73 YGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLV 118
Y ++ P NP W + V GH GC+L+
Sbjct 118 YDEVMRYNPKNPY------WFNRDRFVLSAGH---------GCMLL 148
> At1g31970
Length=537
Score = 28.5 bits (62), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 33/76 (43%), Gaps = 9/76 (11%)
Query 30 PIDTSKWPLLLKNYDRLNV-RTGHYTPLPFGSSPLARPLQQHLNYGSLLLDKPANPS--- 85
PI + WP LL D + + +TG L FG + L+++ G K NP+
Sbjct 139 PIQSHTWPFLLDGRDLIGIAKTGSGKTLAFGIPAIMHVLKKNKKIGG--GSKKVNPTCLV 196
Query 86 ---SHEVVAWLKKILR 98
+ E+ + +LR
Sbjct 197 LSPTRELAVQISDVLR 212
Lambda K H
0.315 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40