bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2085_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
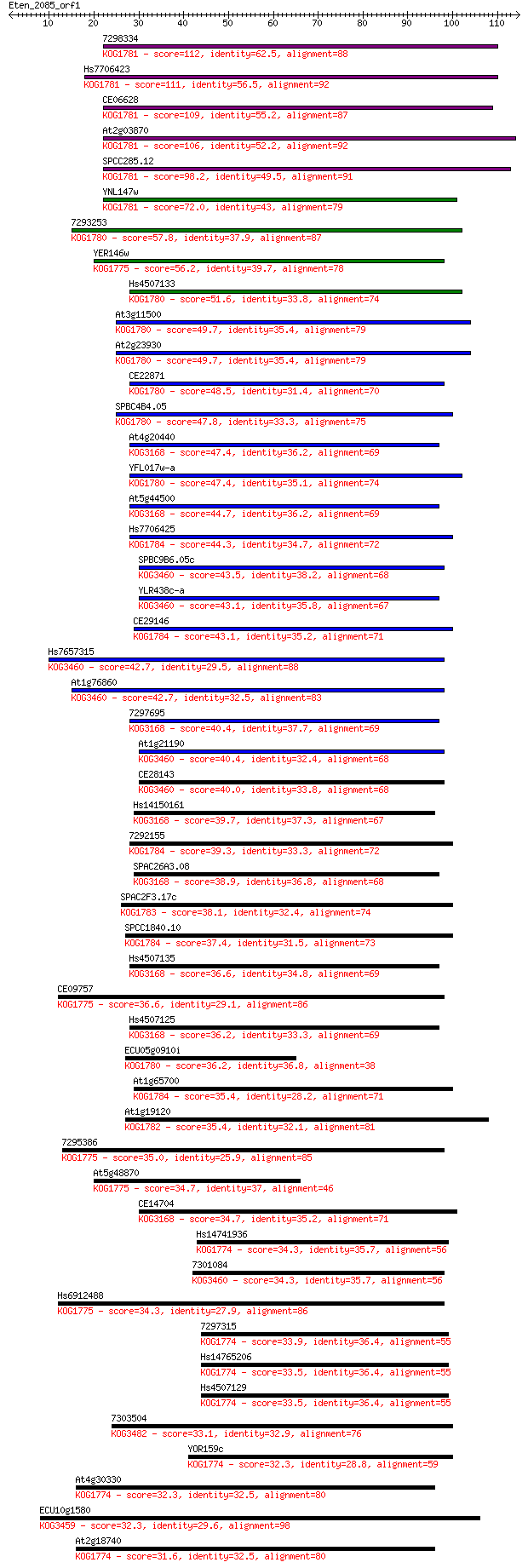
7298334 112 1e-25
Hs7706423 111 3e-25
CE06628 109 1e-24
At2g03870 106 9e-24
SPCC285.12 98.2 4e-21
YNL147w 72.0 3e-13
7293253 57.8 5e-09
YER146w 56.2 1e-08
Hs4507133 51.6 3e-07
At3g11500 49.7 1e-06
At2g23930 49.7 1e-06
CE22871 48.5 3e-06
SPBC4B4.05 47.8 6e-06
At4g20440 47.4 7e-06
YFL017w-a 47.4 7e-06
At5g44500 44.7 4e-05
Hs7706425 44.3 5e-05
SPBC9B6.05c 43.5 1e-04
YLR438c-a 43.1 1e-04
CE29146 43.1 1e-04
Hs7657315 42.7 2e-04
At1g76860 42.7 2e-04
7297695 40.4 8e-04
At1g21190 40.4 8e-04
CE28143 40.0 0.001
Hs14150161 39.7 0.001
7292155 39.3 0.002
SPAC26A3.08 38.9 0.003
SPAC2F3.17c 38.1 0.004
SPCC1840.10 37.4 0.007
Hs4507135 36.6 0.010
CE09757 36.6 0.012
Hs4507125 36.2 0.015
ECU05g0910i 36.2 0.016
At1g65700 35.4 0.026
At1g19120 35.4 0.029
7295386 35.0 0.032
At5g48870 34.7 0.040
CE14704 34.7 0.050
Hs14741936 34.3 0.053
7301084 34.3 0.060
Hs6912488 34.3 0.063
7297315 33.9 0.076
Hs14765206 33.5 0.095
Hs4507129 33.5 0.11
7303504 33.1 0.14
YOR159c 32.3 0.19
At4g30330 32.3 0.21
ECU10g1580 32.3 0.24
At2g18740 31.6 0.33
> 7298334
Length=110
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 55/89 (61%), Positives = 71/89 (79%), Gaps = 1/89 (1%)
Query 22 SVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLL-ETTR 80
S++D SK+L K++RVKF+GGRE G LKG+DA+ NLVLD+T E+LRD ++ +L E TR
Sbjct 20 SILDLSKYLEKQIRVKFAGGREASGILKGYDALLNLVLDNTVEYLRDSDEPYKLTEEQTR 79
Query 81 SLGLVVARGSAIVLISPVDGVQRIENPFI 109
SLGLVV RG+A+VLI P DGV+ I NPFI
Sbjct 80 SLGLVVCRGTALVLICPQDGVESIANPFI 108
> Hs7706423
Length=103
Score = 111 bits (277), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 52/92 (56%), Positives = 68/92 (73%), Gaps = 0/92 (0%)
Query 18 KDPRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLE 77
K S++D SK+++K +RVKF GGRE G LKG D + NLVLD T E++RDP+D +L E
Sbjct 7 KKKESILDLSKYIDKTIRVKFQGGREASGILKGFDPLLNLVLDGTIEYMRDPDDQYKLTE 66
Query 78 TTRSLGLVVARGSAIVLISPVDGVQRIENPFI 109
TR LGLVV RG+++VLI P DG++ I NPFI
Sbjct 67 DTRQLGLVVCRGTSVVLICPQDGMEAIPNPFI 98
> CE06628
Length=104
Score = 109 bits (272), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 48/87 (55%), Positives = 69/87 (79%), Gaps = 0/87 (0%)
Query 22 SVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRS 81
SV+D ++FL+K++RVKF GGRE G L+G D + N+VLDD E+LRDP++ S + + TR
Sbjct 12 SVVDLTRFLDKEIRVKFQGGREASGVLRGFDQLLNMVLDDCREYLRDPQNPSVVGDETRQ 71
Query 82 LGLVVARGSAIVLISPVDGVQRIENPF 108
LGL+VARG+AI ++SP DG+++I NPF
Sbjct 72 LGLIVARGTAITVVSPADGLEQIANPF 98
> At2g03870
Length=99
Score = 106 bits (265), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 70/92 (76%), Gaps = 0/92 (0%)
Query 22 SVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRS 81
+V+D +KF++K V+VK +GGR+V GTLKG+D + NLVLD+ EF+RD +D + + TR
Sbjct 7 TVLDLAKFVDKGVQVKLTGGRQVTGTLKGYDQLLNLVLDEAVEFVRDHDDPLKTTDQTRR 66
Query 82 LGLVVARGSAIVLISPVDGVQRIENPFIGVEA 113
LGL+V RG+A++L+SP DG + I NPF+ EA
Sbjct 67 LGLIVCRGTAVMLVSPTDGTEEIANPFVTAEA 98
> SPCC285.12
Length=113
Score = 98.2 bits (243), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 45/91 (49%), Positives = 66/91 (72%), Gaps = 1/91 (1%)
Query 22 SVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRS 81
S++D S++ +++++ F+GGR++ G LKG D + NLVLDD EE LR+PED +L R
Sbjct 24 SILDLSRYQDQRIQATFTGGRQITGILKGFDQLMNLVLDDVEEQLRNPEDG-KLTGAIRK 82
Query 82 LGLVVARGSAIVLISPVDGVQRIENPFIGVE 112
LGLVV RG+ +VLI+P+DG + I NPF+ E
Sbjct 83 LGLVVVRGTTLVLIAPMDGSEEIPNPFVQAE 113
> YNL147w
Length=107
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 34/82 (41%), Positives = 55/82 (67%), Gaps = 3/82 (3%)
Query 22 SVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASR---LLET 78
+++D +K+ + K+RVK GG+ V G LKG+D + NLVLDDT E++ +P+D + + +
Sbjct 18 AILDLAKYKDSKIRVKLMGGKLVIGVLKGYDQLMNLVLDDTVEYMSNPDDENNTELISKN 77
Query 79 TRSLGLVVARGSAIVLISPVDG 100
R LGL V RG+ +V +S +G
Sbjct 78 ARKLGLTVIRGTILVSLSSAEG 99
> 7293253
Length=76
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 51/87 (58%), Gaps = 11/87 (12%)
Query 15 MSSKDPRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASR 74
MS P V K+++K++ +K +GGR V G L+G D N+VLDDT E +D
Sbjct 1 MSKAHPPEV---KKYMDKRMMLKLNGGRAVTGILRGFDPFMNVVLDDTVEECKD------ 51
Query 75 LLETTRSLGLVVARGSAIVLISPVDGV 101
T ++G+VV RG++IV++ +D V
Sbjct 52 --NTKNNIGMVVIRGNSIVMVEALDRV 76
> YER146w
Length=93
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 45/78 (57%), Gaps = 2/78 (2%)
Query 20 PRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETT 79
P VID K +N+KV + RE EGTL G D N++L+D E+L DPED SR +
Sbjct 8 PLEVID--KTINQKVLIVLQSNREFEGTLVGFDDFVNVILEDAVEWLIDPEDESRNEKVM 65
Query 80 RSLGLVVARGSAIVLISP 97
+ G ++ G+ I ++ P
Sbjct 66 QHHGRMLLSGNNIAILVP 83
> Hs4507133
Length=76
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 45/74 (60%), Gaps = 8/74 (10%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVA 87
KF++KK+ +K +GGR V+G L+G D NLV+D+ E + ++G+VV
Sbjct 11 KFMDKKLSLKLNGGRHVQGILRGFDPFMNLVIDECVEMATSGQQ--------NNIGMVVI 62
Query 88 RGSAIVLISPVDGV 101
RG++I+++ ++ V
Sbjct 63 RGNSIIMLEALERV 76
> At3g11500
Length=79
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 46/79 (58%), Gaps = 9/79 (11%)
Query 25 DPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGL 84
D K+++KK+++K + R V GTL+G D NLV+D+T E D + +G+
Sbjct 9 DLKKYMDKKLQIKLNANRMVVGTLRGFDQFMNLVVDNTVEVNGDDK---------TDIGM 59
Query 85 VVARGSAIVLISPVDGVQR 103
VV RG++IV + ++ V R
Sbjct 60 VVIRGNSIVTVEALEPVGR 78
> At2g23930
Length=80
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 47/79 (59%), Gaps = 9/79 (11%)
Query 25 DPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGL 84
D K+++KK+++K + R V GTL+G D NLV+D+T E + D + +G+
Sbjct 9 DLKKYMDKKLQIKLNANRMVTGTLRGFDQFMNLVVDNTVEV--NGNDKT-------DIGM 59
Query 85 VVARGSAIVLISPVDGVQR 103
VV RG++IV + ++ V R
Sbjct 60 VVIRGNSIVTVEALEPVGR 78
> CE22871
Length=77
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 43/70 (61%), Gaps = 8/70 (11%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVA 87
K+++K++ +K +G R V G L+G D N+V+D+ E+ +D + +LG+ V
Sbjct 11 KYMDKEMDLKLNGNRRVSGILRGFDPFMNMVIDEAVEYQKDG--------GSVNLGMTVI 62
Query 88 RGSAIVLISP 97
RG+++V++ P
Sbjct 63 RGNSVVIMEP 72
> SPBC4B4.05
Length=77
Score = 47.8 bits (112), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 44/75 (58%), Gaps = 8/75 (10%)
Query 25 DPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGL 84
D K+L+++V V+ +G R+V G L+G+D N+VL+D+ E D E +G
Sbjct 8 DLKKYLDRQVFVQLNGSRKVYGVLRGYDIFLNIVLEDSIEEKVDGEKV--------KIGS 59
Query 85 VVARGSAIVLISPVD 99
V RG+++++I +D
Sbjct 60 VAIRGNSVIMIETLD 74
> At4g20440
Length=257
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLL----ETTRSLG 83
+F+N ++RV GR++ G D NLVL D EEF + P + + E R+LG
Sbjct 11 QFINYRMRVTIQDGRQLVGKFMAFDRHMNLVLGDCEEFRKLPPAKGKKINEEREDRRTLG 70
Query 84 LVVARGSAIVLIS 96
LV+ RG ++ ++
Sbjct 71 LVLLRGEEVISMT 83
> YFL017w-a
Length=77
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 44/75 (58%), Gaps = 7/75 (9%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGL-VV 86
K+++KK+ + +G R+V G L+G+D N+VLDD E + ED + LGL V
Sbjct 9 KYMDKKILLNINGSRKVAGILRGYDIFLNVVLDDAMEI--NGEDPA----NNHQLGLQTV 62
Query 87 ARGSAIVLISPVDGV 101
RG++I+ + +D +
Sbjct 63 IRGNSIISLEALDAI 77
> At5g44500
Length=254
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 40/74 (54%), Gaps = 5/74 (6%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDP-----EDASRLLETTRSL 82
+F+N ++RV GR++ G D NLVL D EEF + P + + E R+L
Sbjct 11 QFINYRMRVTIQDGRQLIGKFMAFDRHMNLVLGDCEEFRKLPPAKGNKKTNEEREERRTL 70
Query 83 GLVVARGSAIVLIS 96
GLV+ RG ++ ++
Sbjct 71 GLVLLRGEEVISMT 84
> Hs7706425
Length=96
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 40/77 (51%), Gaps = 14/77 (18%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRS-----L 82
++N+ V V S GR + GTLKG D NL+LD++ E R+ +++ L
Sbjct 7 NYINRTVAVITSDGRMIVGTLKGFDQTINLILDESHE---------RVFSSSQGVEQVVL 57
Query 83 GLVVARGSAIVLISPVD 99
GL + RG + +I +D
Sbjct 58 GLYIVRGDNVAVIGEID 74
> SPBC9B6.05c
Length=93
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 5/73 (6%)
Query 30 LNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEE----FLRDPEDASRLLETTR-SLGL 84
L++ V VK G RE+ G L +D N+VL D EE F + D + L+T R +
Sbjct 18 LDEIVYVKLRGDRELNGRLHAYDEHLNMVLGDAEEIVTIFDDEETDKDKALKTIRKHYEM 77
Query 85 VVARGSAIVLISP 97
+ RG +++LI+P
Sbjct 78 LFVRGDSVILIAP 90
> YLR438c-a
Length=89
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 30 LNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVARG 89
L+++V +K G R + GTL+ D+ CN+VL D E + + L E+ R +V RG
Sbjct 12 LDERVYIKLRGARTLVGTLQAFDSHCNIVLSDAVETIYQLNN-EELSESERRCEMVFIRG 70
Query 90 SAIVLIS 96
+ LIS
Sbjct 71 DTVTLIS 77
> CE29146
Length=98
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 4/71 (5%)
Query 29 FLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVAR 88
++N+ V V GR + G LKG D + NLV++D E R + +L T LGL + R
Sbjct 8 YMNRMVNVVTGDGRVIVGLLKGFDQLINLVIEDAHE--RSYSETEGVL--TTPLGLYIIR 63
Query 89 GSAIVLISPVD 99
G + +I +D
Sbjct 64 GENVAIIGEID 74
> Hs7657315
Length=102
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 51/93 (54%), Gaps = 7/93 (7%)
Query 10 EESTKMSSKDPRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLR-- 67
++ T + ++P +I S L++++ VK RE+ G L +D N++L D EE +
Sbjct 7 QQQTTNTVEEPLDLIRLS--LDERIYVKMRNDRELRGRLHAYDQHLNMILGDVEETVTTI 64
Query 68 --DPEDASRLLETT-RSLGLVVARGSAIVLISP 97
D E + ++T R++ ++ RG +VL++P
Sbjct 65 EIDEETYEEIYKSTKRNIPMLFVRGDGVVLVAP 97
> At1g76860
Length=98
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 47/92 (51%), Gaps = 9/92 (9%)
Query 15 MSSKDPRSVIDPSKF----LNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLR--- 67
MS ++ +V +P L++++ VK RE+ G L D N++L D EE +
Sbjct 1 MSGEEEATVREPLDLIRLSLDERIYVKLRSDRELRGKLHAFDQHLNMILGDVEETITTVE 60
Query 68 -DPEDASRLLETT-RSLGLVVARGSAIVLISP 97
D E ++ TT R++ + RG ++L+SP
Sbjct 61 IDDETYEEIVRTTKRTIEFLFVRGDGVILVSP 92
> 7297695
Length=199
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 36/71 (50%), Gaps = 2/71 (2%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRL--LETTRSLGLV 85
+ LN +VR+ R GT K D NL+L D EEF + S++ E R LG V
Sbjct 11 QHLNYRVRIVLQDSRTFIGTFKAFDKHMNLILGDCEEFRKIRSKNSKVPEREEKRVLGFV 70
Query 86 VARGSAIVLIS 96
+ RG IV ++
Sbjct 71 LLRGENIVSLT 81
> At1g21190
Length=97
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 38/73 (52%), Gaps = 5/73 (6%)
Query 30 LNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLR----DPEDASRLLETT-RSLGL 84
+ +++ VK RE+ G L D N++L D EE + D E ++ TT R++
Sbjct 20 IEERIYVKLRSDRELRGKLHAFDQHLNMILGDVEEVITTIEIDDETYEEIVRTTKRTVPF 79
Query 85 VVARGSAIVLISP 97
+ RG ++L+SP
Sbjct 80 LFVRGDGVILVSP 92
> CE28143
Length=102
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 38/73 (52%), Gaps = 5/73 (6%)
Query 30 LNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLE-----TTRSLGL 84
L+++V VK RE+ G L+ D N+VL + EE + E E T R + +
Sbjct 25 LDERVYVKMRNDRELRGRLRAFDQHLNMVLSEVEETITTREVDEDTFEEIYKQTKRVVPM 84
Query 85 VVARGSAIVLISP 97
+ RG +++L+SP
Sbjct 85 LFVRGDSVILVSP 97
> Hs14150161
Length=125
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 2/67 (2%)
Query 29 FLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVAR 88
LNK +R++ + GR + G D CN++L +EFL+ P D+ E R LGL +
Sbjct 48 LLNKTMRIRMTDGRTLVGCFLCTDRDCNVILGSAQEFLK-PSDSFSAGE-PRVLGLAMVP 105
Query 89 GSAIVLI 95
G IV I
Sbjct 106 GHHIVSI 112
> 7292155
Length=95
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 36/77 (46%), Gaps = 14/77 (18%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRS-----L 82
++N V + + GR GTLKG D N+++D+ E R+ TT L
Sbjct 6 SYINHTVSIITADGRNFIGTLKGFDQTINIIIDECHE---------RVFSTTSGIEQIVL 56
Query 83 GLVVARGSAIVLISPVD 99
GL + RG I +I +D
Sbjct 57 GLHIIRGDNIAVIGLID 73
> SPAC26A3.08
Length=147
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 34/71 (47%), Gaps = 3/71 (4%)
Query 29 FLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPED---ASRLLETTRSLGLV 85
LN + V GR G L D NLVL D +E+ + ++ + E R LGLV
Sbjct 9 LLNHSLNVTTKDGRTFVGQLLAFDGFMNLVLSDCQEYRHIKKQNVPSNSVYEEKRMLGLV 68
Query 86 VARGSAIVLIS 96
+ RG IV +S
Sbjct 69 ILRGEFIVSLS 79
> SPAC2F3.17c
Length=75
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 39/78 (50%), Gaps = 12/78 (15%)
Query 26 PSKFLNK----KVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRS 81
P++FLNK KV ++ S G + +G L D NL L+ TEE++ + T
Sbjct 5 PNEFLNKVIGKKVLIRLSSGVDYKGILSCLDGYMNLALERTEEYVNGKK--------TNV 56
Query 82 LGLVVARGSAIVLISPVD 99
G RG+ ++ +S +D
Sbjct 57 YGDAFIRGNNVLYVSALD 74
> SPCC1840.10
Length=94
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 40/74 (54%), Gaps = 6/74 (8%)
Query 27 SKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDT-EEFLRDPEDASRLLETTRSLGLV 85
+ F+ ++V+V + GR V G+LKG D NL+L D+ E + +D T LG+
Sbjct 4 ADFMEQRVQVITNDGRVVLGSLKGFDHTTNLILSDSFERIISMDQDME-----TIPLGVY 58
Query 86 VARGSAIVLISPVD 99
+ RG + ++ V+
Sbjct 59 LLRGENVAMVGLVN 72
> Hs4507135
Length=240
Score = 36.6 bits (83), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLR-DPEDASRL-LETTRSLGLV 85
+ ++ ++R GR GT K D NL+L D +EF + P++A + E R LGLV
Sbjct 11 QHIDYRMRCILQDGRIFIGTFKAFDKHMNLILCDCDEFRKIKPKNAKQPEREEKRVLGLV 70
Query 86 VARGSAIVLIS 96
+ RG +V ++
Sbjct 71 LLRGENLVSMT 81
> CE09757
Length=91
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 42/86 (48%), Gaps = 7/86 (8%)
Query 12 STKMSSKDPRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPED 71
ST ++ P +ID K + K+ V +E+ GTL G D N+VL+D E+ + D
Sbjct 6 STNPNTLLPLELID--KCIGSKIWVIMKNDKEIVGTLTGFDDYVNMVLEDVVEY-ENTAD 62
Query 72 ASRLLETTRSLGLVVARGSAIVLISP 97
R+ L ++ G+ I ++ P
Sbjct 63 GKRM----TKLDTILLNGNHITMLVP 84
> Hs4507125
Length=231
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Query 28 KFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLR-DPEDASRL-LETTRSLGLV 85
+ ++ ++R GR GT K D NL+L D +EF + P+++ + E R LGLV
Sbjct 11 QHIDYRMRCILQDGRIFIGTFKAFDKHMNLILCDCDEFRKIKPKNSKQAEREEKRVLGLV 70
Query 86 VARGSAIVLIS 96
+ RG +V ++
Sbjct 71 LLRGENLVSMT 81
> ECU05g0910i
Length=78
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 27 SKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEE 64
+ L+K++ V+ GG ++G L G+DA N+VL + E
Sbjct 10 ASLLHKRIVVRIGGGHTLKGLLTGYDAFMNIVLSEVTE 47
> At1g65700
Length=98
Score = 35.4 bits (80), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 37/71 (52%), Gaps = 4/71 (5%)
Query 29 FLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVAR 88
+++ + V + GR + G LKG D N++LD++ E + ++ + LGL + R
Sbjct 10 LVDQIISVITNDGRNIVGVLKGFDQATNIILDESHERVFSTKEGVQ----QHVLGLYIIR 65
Query 89 GSAIVLISPVD 99
G I +I +D
Sbjct 66 GDNIGVIGELD 76
> At1g19120
Length=128
Score = 35.4 bits (80), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 42/81 (51%), Gaps = 6/81 (7%)
Query 27 SKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVV 86
+ +L+KK+ V GR++ G L+ D N VL++ E + + L LGL +
Sbjct 16 AAYLDKKLLVLLRDGRKLMGLLRSFDQFANAVLEEAYERVIVGD-----LYCDIPLGLYI 70
Query 87 ARGSAIVLISPVDGVQRIENP 107
RG +VLI +D V++ E P
Sbjct 71 IRGENVVLIGELD-VEKEELP 90
> 7295386
Length=91
Score = 35.0 bits (79), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 42/85 (49%), Gaps = 7/85 (8%)
Query 13 TKMSSKDPRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDA 72
+ +S+ P ++D K + ++ + +E+ GTL G D N++LDD E+ P D
Sbjct 8 SNISTLMPLELVD--KCIGSRIHIIMKNDKEMVGTLLGFDDFVNMLLDDVTEYENTP-DG 64
Query 73 SRLLETTRSLGLVVARGSAIVLISP 97
R+ L ++ G+ I ++ P
Sbjct 65 RRI----TKLDQILLNGNNITMLVP 85
> At5g48870
Length=88
Score = 34.7 bits (78), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query 20 PRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEF 65
P +ID + + K+ V G +E+ G LKG D N+VL+D E+
Sbjct 10 PSELID--RCIGSKIWVIMKGDKELVGILKGFDVYVNMVLEDVTEY 53
> CE14704
Length=160
Score = 34.7 bits (78), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 37/73 (50%), Gaps = 3/73 (4%)
Query 30 LNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLL--ETTRSLGLVVA 87
LN ++++ GR G K D N++L + EE + A + E R LGLV+
Sbjct 13 LNYRMKIILQDGRTFIGFFKAFDKHMNILLAECEEHRQIKPKAGKKTDGEEKRILGLVLV 72
Query 88 RGSAIVLISPVDG 100
RG IV ++ VDG
Sbjct 73 RGEHIVSMT-VDG 84
> Hs14741936
Length=92
Score = 34.3 bits (77), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 7/56 (12%)
Query 43 EVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVARGSAIVLISPV 98
++EG + G D NLVLDD EE + SR + LG ++ +G I L+ V
Sbjct 42 QIEGCIIGFDEYMNLVLDDAEEI--HSKTKSR-----KQLGRIMLKGDNITLLQSV 90
> 7301084
Length=143
Score = 34.3 bits (77), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 32/61 (52%), Gaps = 5/61 (8%)
Query 42 REVEGTLKGHDAVCNLVLDDTEEFLR----DPEDASRLLETT-RSLGLVVARGSAIVLIS 96
RE+ G L D N+VL D EE + D E + +T R++ ++ RG ++L+S
Sbjct 5 RELRGRLHAFDQHLNMVLGDAEETVTTVEIDEETYEEVYKTAKRTIPMLFVRGDGVILVS 64
Query 97 P 97
P
Sbjct 65 P 65
> Hs6912488
Length=91
Score = 34.3 bits (77), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 41/86 (47%), Gaps = 7/86 (8%)
Query 12 STKMSSKDPRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPED 71
+T S P ++D K + ++ + +E+ GTL G D N+VL+D EF PE
Sbjct 6 TTNPSQLLPLELVD--KCIGSRIHIVMKSDKEIVGTLLGFDDFVNMVLEDVTEFEITPE- 62
Query 72 ASRLLETTRSLGLVVARGSAIVLISP 97
R+ L ++ G+ I ++ P
Sbjct 63 GRRI----TKLDQILLNGNNITMLVP 84
> 7297315
Length=94
Score = 33.9 bits (76), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 26/55 (47%), Gaps = 7/55 (12%)
Query 44 VEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVARGSAIVLISPV 98
+EG + G D NLVLDD EE R+LG ++ +G I LI V
Sbjct 42 IEGHIVGFDEYMNLVLDDAEEVYVKTRQR-------RNLGRIMLKGDNITLIQNV 89
> Hs14765206
Length=92
Score = 33.5 bits (75), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 28/55 (50%), Gaps = 7/55 (12%)
Query 44 VEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVARGSAIVLISPV 98
+EG + G D NLVLDD EE + SR + LG ++ +G I L+ V
Sbjct 43 IEGCIIGFDEYMNLVLDDAEEI--HSKTKSR-----KQLGRIMLKGGNITLLQSV 90
> Hs4507129
Length=92
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 28/55 (50%), Gaps = 7/55 (12%)
Query 44 VEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVARGSAIVLISPV 98
+EG + G D NLVLDD EE + SR + LG ++ +G I L+ V
Sbjct 43 IEGCIIGFDEYMNLVLDDAEEI--HSKTKSR-----KQLGRIMLKGDNITLLQSV 90
> 7303504
Length=84
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 12/80 (15%)
Query 24 IDPSKFLN----KKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETT 79
I+P FLN K V VK G+E +G L D N+ L +TEE + T
Sbjct 3 INPKPFLNGLTGKPVLVKLKWGQEYKGFLVSVDGYMNMQLANTEEVIEG--------SVT 54
Query 80 RSLGLVVARGSAIVLISPVD 99
+LG V+ R + ++ I ++
Sbjct 55 GNLGEVLIRCNNVLYIKGME 74
> YOR159c
Length=94
Score = 32.3 bits (72), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 41 GREVEGTLKGHDAVCNLVLDDTEEFLRDPEDASRLLETTRSLGLVVARGSAIVLISPVD 99
G ++G + G D N+V+D+ E + D +E LG ++ +G I LI+ D
Sbjct 36 GIRIKGKIVGFDEFMNVVIDEAVEIPVNSADGKEDVEKGTPLGKILLKGDNITLITSAD 94
> At4g30330
Length=86
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 41/88 (46%), Gaps = 15/88 (17%)
Query 16 SSKDPRSVIDP----SKFLNKKVRVKFSGGRE----VEGTLKGHDAVCNLVLDDTEEFLR 67
S+K R + P +FL K R++ + +EG + G D NLVLD+ EE
Sbjct 3 STKVQRIMTQPINLIFRFLQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEE--- 59
Query 68 DPEDASRLLETTRSLGLVVARGSAIVLI 95
S +T + LG ++ +G I L+
Sbjct 60 ----VSIKKKTRKPLGRILLKGDNITLM 83
> ECU10g1580
Length=135
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 48/103 (46%), Gaps = 6/103 (5%)
Query 8 PVEESTKMSSKDPRSVIDPSKFLNKKVRVKFSGGREVEGTLKGHDAVCNLVLDDTEEF-- 65
P E+ T M K P S++ + K V V R+V G + +D NL+++D +E
Sbjct 10 PNEKET-MEMKGPLSLVRRAMVKMKPVLVSLRSNRKVLGRVVAYDRHYNLLMEDAKELGT 68
Query 66 LRDPEDASRL---LETTRSLGLVVARGSAIVLISPVDGVQRIE 105
R + E +R LG V RG ++L++ V ++E
Sbjct 69 TRGKNKGRKKRQGCEFSRKLGKVFIRGDTVILVAGEIEVDQVE 111
> At2g18740
Length=88
Score = 31.6 bits (70), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 40/88 (45%), Gaps = 15/88 (17%)
Query 16 SSKDPRSVIDP----SKFLNKKVRVKFSGGRE----VEGTLKGHDAVCNLVLDDTEEFLR 67
S+K R + P +FL K R++ + +EG + G D NLVLD+ EE
Sbjct 3 STKVQRIMTQPINLIFRFLQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEE--- 59
Query 68 DPEDASRLLETTRSLGLVVARGSAIVLI 95
S T + LG ++ +G I L+
Sbjct 60 ----VSIKKNTRKPLGRILLKGDNITLM 83
Lambda K H
0.316 0.136 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181971906
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40