bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2121_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
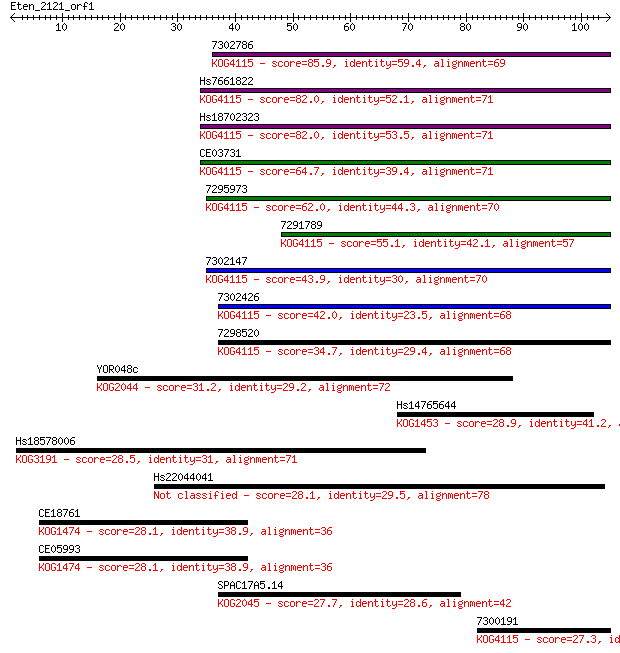
7302786 85.9 2e-17
Hs7661822 82.0 3e-16
Hs18702323 82.0 3e-16
CE03731 64.7 4e-11
7295973 62.0 3e-10
7291789 55.1 3e-08
7302147 43.9 8e-05
7302426 42.0 3e-04
7298520 34.7 0.040
YOR048c 31.2 0.42
Hs14765644 28.9 2.6
Hs18578006 28.5 3.0
Hs22044041 28.1 3.6
CE18761 28.1 3.8
CE05993 28.1 3.8
SPAC17A5.14 27.7 5.2
7300191 27.3 6.9
> 7302786
Length=97
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/69 (59%), Positives = 51/69 (73%), Gaps = 0/69 (0%)
Query 36 EVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDLDP 95
EVEETL RI++ KGV G IV+ +G ++STL T YA +QLAD+ARS+VRDLDP
Sbjct 4 EVEETLKRIQSHKGVVGTIVVNNEGIPVKSTLDNTTTVQYAGLMSQLADKARSVVRDLDP 63
Query 96 QNDVTFLRV 104
ND+TFLRV
Sbjct 64 SNDMTFLRV 72
> Hs7661822
Length=96
Score = 82.0 bits (201), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/71 (52%), Positives = 52/71 (73%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVEETL R++++KGV+GIIV+ +G ++ST+ T YA+ +ARS VRD+
Sbjct 1 MAEVEETLKRLQSQKGVQGIIVVNTEGIPIKSTMDNPTTTQYASLMHSFILKARSTVRDI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> Hs18702323
Length=96
Score = 82.0 bits (201), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 50/71 (70%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVEETL RI++ KGV G +V+ A+G +R+TL T YA L +A+S VRD+
Sbjct 1 MAEVEETLKRIQSHKGVIGTMVVNAEGIPIRTTLDNSTTVQYAGLLHHLTMKAKSTVRDI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> CE03731
Length=95
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 49/71 (69%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
M++ EET+ R+++ KGV GIIV+ + G ++ ST+ T ++ A QL ++ ++ +R+L
Sbjct 1 MSDFEETIRRLQSEKGVVGIIVVDSAGRVIHSTIDSDATQSHTAFLQQLCEKTKTSIREL 60
Query 94 DPQNDVTFLRV 104
D ND+TFLR+
Sbjct 61 DSSNDLTFLRL 71
> 7295973
Length=97
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 44/70 (62%), Gaps = 0/70 (0%)
Query 35 AEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDLD 94
AEVEE L R ++ K V GI+V+ DG +++TL T YAA + ++AR +V DLD
Sbjct 3 AEVEELLKRFQSMKNVTGIVVVDNDGIPIKTTLDYTLTLHYAALMQTVREKARQVVLDLD 62
Query 95 PQNDVTFLRV 104
N+ TFLR+
Sbjct 63 ATNEFTFLRL 72
> 7291789
Length=130
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 24/57 (42%), Positives = 38/57 (66%), Gaps = 0/57 (0%)
Query 48 KGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDLDPQNDVTFLRV 104
+G + I++L + G LRST Q++T + + L AR++VRDLDP ND+TF+R+
Sbjct 42 RGARDIMILDSHGVPLRSTCSQRRTFVFVSNLKPLLFMARNVVRDLDPSNDITFMRI 98
> 7302147
Length=97
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 38/70 (54%), Gaps = 0/70 (0%)
Query 35 AEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDLD 94
AE+E+ L R + V GII+L +++T+ T YAA + L +A ++ +LD
Sbjct 3 AEIEDLLKRYQNYPNVSGIIILDPFAIPIKTTMEYTLTVHYAALISTLTYKAAKMITNLD 62
Query 95 PQNDVTFLRV 104
N++ +R+
Sbjct 63 ASNELVTIRL 72
> 7302426
Length=114
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 16/68 (23%), Positives = 40/68 (58%), Gaps = 0/68 (0%)
Query 37 VEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDLDPQ 96
VEE +++ + GV+ I+++ G +++++ +Q+ YA L ++ ++ + ++P
Sbjct 15 VEEVFRKVQEKPGVEDILIMNHSGVPVKTSMDRQEGLQYACLYDNLREKCQAFLSKMEPA 74
Query 97 NDVTFLRV 104
++T LRV
Sbjct 75 QNLTLLRV 82
> 7298520
Length=115
Score = 34.7 bits (78), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 38/68 (55%), Gaps = 0/68 (0%)
Query 37 VEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDLDPQ 96
V+ T +R+ GV G I++ +G +R+ L YA L ARS+V+DL+
Sbjct 6 VQMTFDRLVQLPGVTGAILIDGNGVPVRTNLPANVARIYADRMRPLVILARSMVQDLENG 65
Query 97 NDVTFLRV 104
++++++R+
Sbjct 66 DELSYVRL 73
> YOR048c
Length=1006
Score = 31.2 bits (69), Expect = 0.42, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Query 16 PCCFLRSVTPVFAEASKKMAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAY 75
PC + P E +A E T NR+ + ++V+A DG R+ + QQ+ +
Sbjct 60 PCSHPENKPPPETEDEMLLAVFEYT-NRVLNMARPRKVLVMAVDGVAPRAKMNQQRARRF 118
Query 76 AAAA-AQLADRAR 87
+A AQ+ + AR
Sbjct 119 RSARDAQIENEAR 131
> Hs14765644
Length=1136
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 68 GQQQTAAYAAAAAQLADRARSLVRDLDPQNDVTF 101
G+Q + A A LA R R L+RDL P+N +
Sbjct 873 GRQDGSESEAVAVALAGRLRELLRDLPPENRASL 906
> Hs18578006
Length=174
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 35/78 (44%), Gaps = 14/78 (17%)
Query 2 PQTLNPRVWLGPSTPCCFLRS-------VTPVFAEASKKMAEVEETLNRIKTRKGVKGII 54
PQ L + P C L++ + PV + +AE E++L R R+G +
Sbjct 52 PQALYMCTDINPEAAACTLKTARCNKVHIQPVITDVQSVLAETEKSLQR---RRGFE--- 105
Query 55 VLAADGCILRSTLGQQQT 72
LA+ GCI + Q+ T
Sbjct 106 -LASPGCIGFEQMNQKYT 122
> Hs22044041
Length=2375
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 41/79 (51%), Gaps = 6/79 (7%)
Query 26 VFAEASKKMAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADR 85
VF ++ K++AE + LNR R+ + +++ GC+L + + TA A+ L
Sbjct 1347 VFLKSRKRLAEAADLLNREGRRE--EAALLMKQHGCLLEAA---RLTADKDFQASCLLGA 1401
Query 86 AR-SLVRDLDPQNDVTFLR 103
AR ++ RD D ++ LR
Sbjct 1402 ARLNVARDSDIEHTKDILR 1420
> CE18761
Length=1087
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 6 NPRVWLGPSTPCCFLRSVTPVFAEASKKMAEVEETL 41
+P+ G +TPC +PV E + KM EVEE +
Sbjct 153 SPQKPQGSTTPCSVQSPASPVGDEQAVKMEEVEEPI 188
> CE05993
Length=1250
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 6 NPRVWLGPSTPCCFLRSVTPVFAEASKKMAEVEETL 41
+P+ G +TPC +PV E + KM EVEE +
Sbjct 153 SPQKPQGSTTPCSVQSPASPVGDEQAVKMEEVEEPI 188
> SPAC17A5.14
Length=1328
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 12/42 (28%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query 37 VEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAA 78
+E +IK +K ++ +A DGC R+ + QQ++ + A
Sbjct 68 IEHLFEKIKPKK----LLYMAVDGCAPRAKMNQQRSRRFRTA 105
> 7300191
Length=101
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 10/23 (43%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 82 LADRARSLVRDLDPQNDVTFLRV 104
L +S+VRD+DP N + F+R+
Sbjct 58 LVSTCQSVVRDIDPSNKLCFMRL 80
Lambda K H
0.320 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40