bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2148_orf1
Length=340
Score E
Sequences producing significant alignments: (Bits) Value
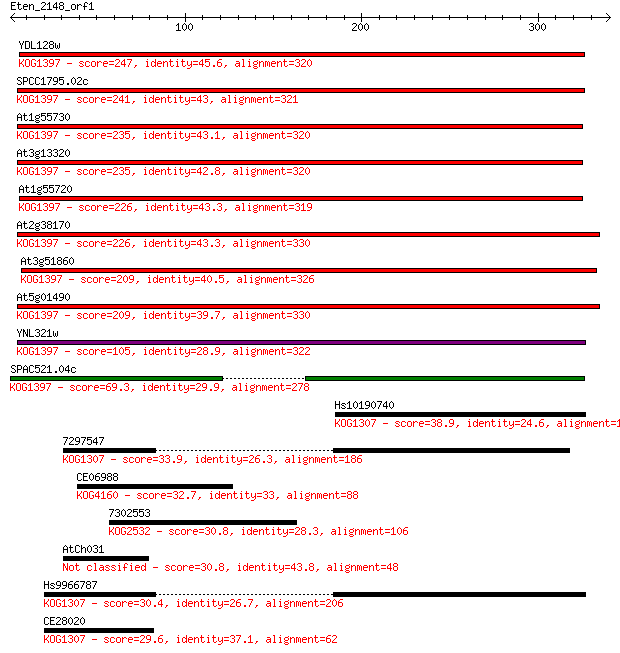
YDL128w 247 3e-65
SPCC1795.02c 241 1e-63
At1g55730 235 8e-62
At3g13320 235 8e-62
At1g55720 226 6e-59
At2g38170 226 8e-59
At3g51860 209 5e-54
At5g01490 209 5e-54
YNL321w 105 2e-22
SPAC521.04c 69.3 1e-11
Hs10190740 38.9 0.018
7297547 33.9 0.51
CE06988 32.7 1.3
7302553 30.8 4.2
AtCh031 30.8 4.8
Hs9966787 30.4 5.2
CE28020 29.6 9.5
> YDL128w
Length=411
Score = 247 bits (630), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 146/334 (43%), Positives = 227/334 (67%), Gaps = 14/334 (4%)
Query 6 FSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVK 65
F FNF A+IPL+A+L TEEL+ G GGLLNATFGNAVE+I+S+ AL+ G + +V+
Sbjct 64 FLFNFLAIIPLAAILANATEELADKAGNTIGGLLNATFGNAVELIVSIIALKKGQVRIVQ 123
Query 66 GTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTV--AAV 123
++LGS+LSNLLLVLG+ F GG + Q FN+ A +LL ++C ++IP A +
Sbjct 124 ASMLGSLLSNLLLVLGLCFIFGGYNRVQQTFNQTAAQTMSSLLAIACASLLIPAAFRATL 183
Query 124 DNGQHGTY---NILMISRITALLIGVTYCLFLFFQLYTHIGLFKDDDEDAEQ-------- 172
+G+ + IL +SR T+++I + Y LFL+FQL +H LF+ +E+ ++
Sbjct 184 PHGKEDHFIDGKILELSRGTSIVILIVYVLFLYFQLGSHHALFEQQEEETDEVMSTISRN 243
Query 173 -WPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEH 231
+S +++ ++L T +++ ++ LV +I+ VV GLS++FIG+I++PIVGNAAEH
Sbjct 244 PHHSLSVKSSLVILLGTTVIISFCADFLVGTIDNVVESTGLSKTFIGLIVIPIVGNAAEH 303
Query 232 LTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLS 291
+T+V VAMK+K+DLA+GVA+GSS Q+ALFV PF VLV W++ P+T+ + ++
Sbjct 304 VTSVLVAMKDKMDLALGVAIGSSLQVALFVTPFMVLVGWMIDVPMTLNFSTFETATLFIA 363
Query 292 VLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWF 325
V ++ ++ DGESNWLEGVM +A Y++I++ F++
Sbjct 364 VFLSNYLILDGESNWLEGVMSLAMYILIAMAFFY 397
> SPCC1795.02c
Length=412
Score = 241 bits (616), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 138/325 (42%), Positives = 216/325 (66%), Gaps = 4/325 (1%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
VF N A+IPL++LL TE+LS+ +G G LLNA+FGNA+E+I+ V AL+ G L +V
Sbjct 85 VFILNMLAIIPLASLLSFATEQLSIISGPTLGALLNASFGNAIELIVGVLALKRGELRIV 144
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTV--AA 122
+ +LLGSILSNLLLV GM G+ + FN A + +L LS I+IP +
Sbjct 145 QSSLLGSILSNLLLVFGMCLVTTGIRREITTFNITVAQTMIAMLALSTATILIPATFHYS 204
Query 123 VDNGQHGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLFKDDDE-DAEQWP-MMSWEA 180
+ + + +L +SR TA+++ + Y L L FQL TH + D E + E P ++ +
Sbjct 205 LPDNANSENALLHVSRGTAVIVLIVYVLLLVFQLKTHKHVCHDPSEVEEETEPRILGLRS 264
Query 181 ATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMK 240
+ ML +VT V+L ++ LV SI+++V + +S++F+G+++LP+VGNAAEH+TA+ V+ +
Sbjct 265 SIAMLAIVTVFVSLCADYLVGSIDQLVEEVNISKTFVGLVILPVVGNAAEHVTAIVVSYR 324
Query 241 NKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVVQ 300
++DLA+GVA+GSS QIALF+ PF V+V W++ QPLT+ + L V++ +SV + ++Q
Sbjct 325 GQMDLALGVAIGSSIQIALFLAPFLVIVGWIISQPLTLYFESLETVILFVSVFLVNYLIQ 384
Query 301 DGESNWLEGVMLMAAYLIISVVFWF 325
DG ++WLEGV L+A Y I+ + F++
Sbjct 385 DGATHWLEGVQLLALYAIVVLAFFY 409
> At1g55730
Length=424
Score = 235 bits (600), Expect = 8e-62, Method: Compositional matrix adjust.
Identities = 138/336 (41%), Positives = 207/336 (61%), Gaps = 20/336 (5%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F + + PL+ LG TE+L+ +TG GGLLNATFGN E+I+S+ AL+ G++ VV
Sbjct 85 IFLLSLVGITPLAERLGYATEQLACYTGSTVGGLLNATFGNVTELIISIFALKSGMIRVV 144
Query 65 KGTLLGSILSNLLLVLGMSFFAGGL--HHYLQKFNEKGATCSVTLLLLSCMGIVIPTVAA 122
+ TLLGSILSN+LLVLG +FF GGL Q F++ A + LLL++ MG++ P V
Sbjct 145 QLTLLGSILSNMLLVLGCAFFCGGLVFSQKEQVFDKGNAVVNSGLLLMAVMGLLFPAVLH 204
Query 123 VDNGQ-HGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLF-------------KDDDE 168
+ + H + L +SR ++ ++ V Y +LFFQL + + DDDE
Sbjct 205 YTHSEVHAGSSELALSRFSSCIMLVAYAAYLFFQLKSQPSSYTPLTEETNQNEETSDDDE 264
Query 169 DAEQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNA 228
D E + WEA + L ++T+ V+L S LV +IE + + SFI VILLPIVGNA
Sbjct 265 DPE---ISKWEA-IIWLSILTAWVSLLSGYLVDAIEGASVSWKIPISFISVILLPIVGNA 320
Query 229 AEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVM 288
AEH A+ AMK+K+DL++GVA+GSS QI++F PF V++ W+MG + + Q +
Sbjct 321 AEHAGAIMFAMKDKLDLSLGVAIGSSIQISMFAVPFCVVIGWMMGAQMDLNFQLFETATL 380
Query 289 LLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFW 324
++V+V +Q+G SN+ +G+ML+ YLI++ F+
Sbjct 381 FITVIVVAFFLQEGTSNYFKGLMLILCYLIVAASFF 416
> At3g13320
Length=439
Score = 235 bits (600), Expect = 8e-62, Method: Compositional matrix adjust.
Identities = 137/332 (41%), Positives = 208/332 (62%), Gaps = 12/332 (3%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
VF + PL+ LG TE+L+ +TG GGLLNATFGN E+I+S+ AL+ G++ VV
Sbjct 100 VFLLTLVGITPLAERLGYATEQLACYTGPTVGGLLNATFGNVTELIISIFALKNGMIRVV 159
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSVTLLLLSCMGIVIPTVAA 122
+ TLLGSILSN+LLVLG +FF GGL Y Q F++ AT + LLL++ MGI+ P V
Sbjct 160 QLTLLGSILSNMLLVLGCAFFCGGLVFYQKDQVFDKGIATVNSGLLLMAVMGILFPAVLH 219
Query 123 VDNGQ-HGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLFKDDDEDAEQW-------- 173
+ + H + L +SR ++ ++ + Y +LFFQL + + DE++ Q
Sbjct 220 YTHSEVHAGSSELALSRFSSCIMLIAYAAYLFFQLKSQSNSYSPLDEESNQNEETSAEDE 279
Query 174 -PMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHL 232
P +S A + L ++T+ V+L S LV +IE + + +FI ILLPIVGNAAEH
Sbjct 280 DPEISKWEAIIWLSILTAWVSLLSGYLVDAIEGASVSWNIPIAFISTILLPIVGNAAEHA 339
Query 233 TAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSV 292
A+ AMK+K+DL++GVA+GSS QI++F PF V++ W+MGQ + + Q ++ ++V
Sbjct 340 GAIMFAMKDKLDLSLGVAIGSSIQISMFAVPFCVVIGWMMGQQMDLNFQLFETAMLFITV 399
Query 293 LVAMAVVQDGESNWLEGVMLMAAYLIISVVFW 324
+V +Q+G SN+ +G+ML+ YLI++ F+
Sbjct 400 IVVAFFLQEGSSNYFKGLMLILCYLIVAASFF 431
> At1g55720
Length=401
Score = 226 bits (576), Expect = 6e-59, Method: Compositional matrix adjust.
Identities = 138/343 (40%), Positives = 207/343 (60%), Gaps = 28/343 (8%)
Query 6 FSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVK 65
F + + PL+ LG TE+LS +TG GGLLNATFGN +E+I+S+ AL+ G++ VV+
Sbjct 55 FLLSLVGITPLAERLGYATEQLSCYTGATVGGLLNATFGNVIELIISIIALKNGMIRVVQ 114
Query 66 GTLLGSILSNLLLVLGMSFFAGGL--HHYLQKFNEKGATCSVTLLLLSCMGIVIPTVAAV 123
TLLGSILSN+LLVLG +FF GGL Q F+++ A S +LL++ MG++ PT
Sbjct 115 LTLLGSILSNILLVLGCAFFCGGLVFPGKDQVFDKRNAVVSSGMLLMAVMGLLFPTFLHY 174
Query 124 DNGQ-HGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLF----------KDDDEDAEQ 172
+ + H + L +SR + ++ V Y +LFFQL + + +DDED E
Sbjct 175 THSEVHAGSSELALSRFISCIMLVAYAAYLFFQLKSQPSFYTEKTNQNEETSNDDEDPE- 233
Query 173 WPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHL 232
+ WEA + L + T+ V+L S LV +IE + + SFI VILLPIVGNAAEH
Sbjct 234 --ISKWEA-IIWLSIFTAWVSLLSGYLVDAIEGTSVSWKIPISFISVILLPIVGNAAEHA 290
Query 233 TAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSV 292
A+ AMK+K+DL++GVA+GSS QI++F PF V++ W+MG + + +Q +L++V
Sbjct 291 GAIMFAMKDKLDLSLGVAIGSSIQISMFAVPFCVVIGWMMGAQMDLNLQLFETATLLITV 350
Query 293 LVAMAVVQ-----------DGESNWLEGVMLMAAYLIISVVFW 324
+V +Q +G SN+ + +ML+ YLI++ F+
Sbjct 351 IVVAFFLQLVNLEANYETNEGTSNYFKRLMLILCYLIVAASFF 393
> At2g38170
Length=463
Score = 226 bits (575), Expect = 8e-59, Method: Compositional matrix adjust.
Identities = 143/351 (40%), Positives = 218/351 (62%), Gaps = 27/351 (7%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F + L PL+ + TE+L+ +TG GGLLNAT GNA E+I+++ AL + VV
Sbjct 98 IFGLSLLGLTPLAERVSFLTEQLAFYTGPTLGGLLNATCGNATELIIAILALTNNKVAVV 157
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSVTLLLLSCMGIVIPT-VA 121
K +LLGSILSNLLLVLG S F GG+ + Q+F+ K A + LLLL + ++P V
Sbjct 158 KYSLLGSILSNLLLVLGTSLFCGGIANIRREQRFDRKQADVNFFLLLLGFLCHLLPLLVG 217
Query 122 AVDNGQHGTYNI----LMISRITALLIGVTYCLFLFFQLYTHIGLF----KDD--DEDAE 171
+ NG+ + L ISR ++++ ++Y +L FQL+TH LF ++D D+D E
Sbjct 218 YLKNGEASAAVLSDMQLSISRGFSIVMLISYIAYLVFQLWTHRQLFDAQEQEDEYDDDVE 277
Query 172 Q-------WPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPI 224
Q W +W L +T ++AL SE +V++IEE + LS SFI +ILLPI
Sbjct 278 QETAVISFWSGFAW------LVGMTLVIALLSEYVVATIEEASDKWNLSVSFISIILLPI 331
Query 225 VGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLS 284
VGNAAEH AV A KNK+D+++GVA+GS+ QI LFV P T++VAW++G + + PL
Sbjct 332 VGNAAEHAGAVIFAFKNKLDISLGVALGSATQIGLFVVPLTIIVAWILGINMDLNFGPLE 391
Query 285 AVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWFDK-PGPESAV 334
+ +S+++ +QDG S++++G++L+ Y II++ F+ DK P ++A+
Sbjct 392 TGCLAVSIIITAFTLQDGSSHYMKGLVLLLCYFIIAICFFVDKLPQKQNAI 442
> At3g51860
Length=448
Score = 209 bits (533), Expect = 5e-54, Method: Compositional matrix adjust.
Identities = 132/343 (38%), Positives = 210/343 (61%), Gaps = 18/343 (5%)
Query 7 SFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVKG 66
S+N+ L PL+ + TE+L+ +TG GGLLNAT GNA E+I+++ AL + VVK
Sbjct 90 SYNYG-LTPLAERVSFLTEQLAFYTGPTVGGLLNATCGNATELIIAILALANNKVAVVKY 148
Query 67 TLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSV-TLLLLSCMGIVIPTVAAV 123
+LLGSILSNLLLVLG S F GG+ + Q+F+ K A + LL+ ++ +
Sbjct 149 SLLGSILSNLLLVLGTSLFFGGIANIRREQRFDRKQADVNFFLLLMGLLCHLLPLLLKYA 208
Query 124 DNGQHGTYNI----LMISRITALLIGVTYCLFLFFQLYTHIGLFKDD---------DEDA 170
G+ T I L +SR +++++ + Y +L FQL+TH LF+ +
Sbjct 209 ATGEVSTSMINKMSLTLSRTSSIVMLIAYIAYLIFQLWTHRQLFEAQQDDDDAYDDEVSV 268
Query 171 EQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAE 230
E+ P++ + + L +T ++AL SE +V +IE+ +GLS SFI +ILLPIVGNAAE
Sbjct 269 EETPVIGFWSGFAWLVGMTIVIALLSEYVVDTIEDASDSWGLSVSFISIILLPIVGNAAE 328
Query 231 HLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLL 290
H A+ A KNK+D+++GVA+GS+ QI+LFV P +V+VAW++G + + L + L
Sbjct 329 HAGAIIFAFKNKLDISLGVALGSATQISLFVVPLSVIVAWILGIKMDLNFNILETSSLAL 388
Query 291 SVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWFDK-PGPES 332
++++ +QDG S++++G++L+ Y+II+ F+ D+ P P
Sbjct 389 AIIITAFTLQDGTSHYMKGLVLLLCYVIIAACFFVDQIPQPND 431
> At5g01490
Length=442
Score = 209 bits (533), Expect = 5e-54, Method: Compositional matrix adjust.
Identities = 131/345 (37%), Positives = 202/345 (58%), Gaps = 18/345 (5%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
VF+ + L PL+ + TE+++ HTG GL+NAT GNA EMI+++ A+ + +V
Sbjct 99 VFALSLLGLTPLAERISFLTEQIAFHTGPT--GLMNATCGNATEMIIAILAVGQRKMRIV 156
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSVTLLLLSCMGIVIPTVAA 122
K +LLGSILSNLL VLG S F GG+ + Q F+ + L L C + +
Sbjct 157 KLSLLGSILSNLLFVLGTSLFLGGISNLRKHQSFDPGDMNSMLLYLALLCQTLPMIMRFT 216
Query 123 VDNGQHGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLF-----------KDDD--ED 169
++ ++ +++++SR ++ ++ + Y FL F L++ DDD +
Sbjct 217 MEAEEYDGSDVVVLSRASSFVMLIAYLAFLIFHLFSSHLSPPPPPLPQREDVHDDDVSDK 276
Query 170 AEQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAA 229
E+ ++ +A L ++T LVAL S+ LVS+I++ +GLS FIG+ILLPIVGNAA
Sbjct 277 EEEGAVIGMWSAIFWLIIMTLLVALLSDYLVSTIQDAADSWGLSVGFIGIILLPIVGNAA 336
Query 230 EHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVML 289
EH AV A +NK+D+ +G+A+GS+ QIALFV P TVLVAW MG + + L
Sbjct 337 EHAGAVIFAFRNKLDITLGIALGSATQIALFVVPVTVLVAWTMGIEMDLNFNLLETACFA 396
Query 290 LSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWFDKPGPESAV 334
LS+LV V+QDG SN+++G++L+ Y++I+ F F P S +
Sbjct 397 LSILVTSLVLQDGTSNYMKGLVLLLCYVVIAACF-FVSNSPSSKL 440
> YNL321w
Length=908
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 93/373 (24%), Positives = 170/373 (45%), Gaps = 51/373 (13%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F + IPL+ +G +S T G ++NA F VE+ + AL+ +V
Sbjct 531 IFILCLTSTIPLAFYIGQAVASISAQTSMGVGAVINAFFSTIVEIFLYCVALQQKKGLLV 590
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTV---- 120
+G+++GSIL +LL+ G+S G L+ Q++N A S LL+ S + + +PTV
Sbjct 591 EGSMIGSILGAVLLLPGLSMCGGALNRKTQRYNPASAGVSSALLIFSMIVMFVPTVLYEI 650
Query 121 ----------AAVDNGQHGTYNILMISRI-----------TALLIGVTYCLFLFFQLYTH 159
A D ++ L +R+ A+++ Y + L+F L TH
Sbjct 651 YGGYSVNCADGANDRDCTFSHPPLKFNRLFTHVIQPMSISCAIVLFCAYIIGLWFTLRTH 710
Query 160 IGLF-------------KDDDEDAEQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEV 206
+ + ++++ P S +T +L + T L A+ +E+LVS ++ V
Sbjct 711 AKMIWQLPIADPTSTAPEQQEQNSHDAPNWSRSKSTCILLMSTLLYAIIAEILVSCVDAV 770
Query 207 VADY-GLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFP-- 263
+ D L+ F+G+ + ++ N E L A++ A+ V L+M + + Q+ L P
Sbjct 771 LEDIPSLNPKFLGLTIFALIPNTTEFLNAISFAIHGNVALSMEIGSAYALQVCLLQIPSL 830
Query 264 --FTVLVAWVMGQPL-TMAVQPLSAVVMLLSVLVAMAVV-------QDGESNWLEGVMLM 313
+++ W + + + + Q V + AM V +G+SN+ +G ML+
Sbjct 831 VIYSIFYTWNVKKSMINIRTQMFPLVFPRWDIFGAMTSVFMFTYLYAEGKSNYFKGSMLI 890
Query 314 AAYLIISVVFWFD 326
Y+II V F+F
Sbjct 891 LLYIIIVVGFYFQ 903
> SPAC521.04c
Length=881
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 71/120 (59%), Gaps = 0/120 (0%)
Query 1 STLCVFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGL 60
S++ +F+ + ++IPL+ +G +S + G +NA FG+ +E+ + ALR G
Sbjct 436 SSVFLFTASLVSIIPLAYFIGMAVASISAQSSMGMGAFINAFFGSVIEVFLYSVALRKGN 495
Query 61 LEVVKGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTV 120
+V+G+++GSIL+ LLL+ G+S AG + Q FN K A + T+LL + +G PT+
Sbjct 496 AGLVEGSVIGSILAGLLLMPGLSMCAGAIRKKFQFFNIKSAGATSTMLLFAVLGAFAPTM 555
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/168 (25%), Positives = 84/168 (50%), Gaps = 13/168 (7%)
Query 168 EDAEQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGN 227
+DA W S + ++L T L +L +E+LV ++ V+ + +SE F+G+ L +V N
Sbjct 714 DDAPNW---SRSKSAIILLSATFLYSLIAEILVEHVDTVLDKFAISEKFLGLTLFALVPN 770
Query 228 AAEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFP----FTVLVAWVMGQPL------T 277
E + A++ A+ + L+M + + Q+ L P +++ + G + T
Sbjct 771 TTEFMNAISFALNENIALSMEIGSAYALQVCLLQIPCLMGYSLFQYYRSGDSISFKHLFT 830
Query 278 MAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWF 325
M + +++ V + V +G+SN+ +G +L+ AYL+ + F F
Sbjct 831 MVFPTWDMICVMICVFLLTYVHSEGKSNYFKGSILVLAYLVSMLGFTF 878
> Hs10190740
Length=593
Score = 38.9 bits (89), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 35/144 (24%), Positives = 67/144 (46%), Gaps = 5/144 (3%)
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
+++ +L + + V S+E++ LSE G + +A E T+V K D
Sbjct 67 IYMFYALAIVCDDFFVPSLEKICERLHLSEDVAGATFMAAGSSAPELFTSVIGVFITKGD 126
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPL--SAVVMLLSVLVAMAVVQDG 302
+ +G VGS+ L + L A GQ + ++ L ++ LSV+ + + D
Sbjct 127 VGVGTIVGSAVFNILCIIGVCGLFA---GQVVALSSWCLLRDSIYYTLSVIALIVFIYDE 183
Query 303 ESNWLEGVMLMAAYLIISVVFWFD 326
+ +W E ++L+ YLI V+ ++
Sbjct 184 KVSWWESLVLVLMYLIYIVIMKYN 207
> 7297547
Length=856
Score = 33.9 bits (76), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 31/136 (22%), Positives = 62/136 (45%), Gaps = 5/136 (3%)
Query 184 MLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKV 243
++++ +L + E V S++ ++ G+++ G + G+A E T+V +
Sbjct 340 VIYMFVALAIVCDEFFVPSLDVIIEKLGITDDVAGATFMAAGGSAPELFTSVIGVFVSFD 399
Query 244 DLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPL--SAVVMLLSVLVAMAVVQD 301
D+ +G VGS+ LFV L + + L++ PL +S+LV + +D
Sbjct 400 DVGIGTIVGSAVFNILFVIGMCALFSKTV---LSLTWWPLFRDCSFYSISLLVLIYFFRD 456
Query 302 GESNWLEGVMLMAAYL 317
W E ++L Y+
Sbjct 457 NRIFWWEALILFTIYI 472
Score = 32.3 bits (72), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 31 TGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLVLGM 82
T ++ G A G+A E+ SV + + +V GT++GS + N+L V+GM
Sbjct 369 TDDVAGATFMAAGGSAPELFTSVIGVFVSFDDVGIGTIVGSAVFNILFVIGM 420
> CE06988
Length=606
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 43/92 (46%), Gaps = 12/92 (13%)
Query 39 LNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLVLGMS----FFAGGLHHYLQ 94
LN+T N + M Q + IGLL G L GS+ N+++ L ++ A GL +L+
Sbjct 108 LNSTAPNKISWTM--QNMDIGLL----GDLSGSV--NIVVPLNLTGQVEILAQGLTFHLE 159
Query 95 KFNEKGATCSVTLLLLSCMGIVIPTVAAVDNG 126
EKG S + LSC+ + NG
Sbjct 160 SSIEKGKNGSAKVTSLSCLATIRDVTVTNHNG 191
> 7302553
Length=462
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 54/106 (50%), Gaps = 5/106 (4%)
Query 57 RIGLLEVVKGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIV 116
+I +++ KG LL S+ ++L+L SFF L LQK ++G + S + + +G
Sbjct 270 KIYHVDIKKGALLSSLPYMVMLLL--SFFFVWLSKVLQK--KEGMSLSFNRKIFNSIGHW 325
Query 117 IPTVAAVDNGQHGTYNILMISRITALLIGVTYCLFLFFQLYTHIGL 162
IP ++ + G N + + L +G++ +L FQ+ HI L
Sbjct 326 IPMLSLIALGYVPADNAPLAVTLLTLTVGISGATYLGFQV-NHIDL 370
> AtCh031
Length=479
Score = 30.8 bits (68), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 31 TGEITGGLLNATFGNAVEMI-MSVQALRIGLLEVVKGTLLGSILSNLLL 78
TGEI G LNAT G EMI +V A +G+ V+ L G +N L
Sbjct 232 TGEIKGHYLNATAGTCEEMIKRAVFARELGVPIVMHDYLTGGFTANTSL 280
> Hs9966787
Length=661
Score = 30.4 bits (67), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 34/145 (23%), Positives = 61/145 (42%), Gaps = 5/145 (3%)
Query 184 MLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKV 243
M+++ +L + E V S+ + G+S+ G + G+A E T++
Sbjct 142 MIYMFIALAIVCDEFFVPSLTVITEKLGISDDVAGATFMAAGGSAPELFTSLIGVFIAHS 201
Query 244 DLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVV--QD 301
++ +G VGS+ LFV L + + L + PL V V + M ++ D
Sbjct 202 NVGIGTIVGSAVFNILFVIGMCALFS---REILNLTWWPLFRDVSFYIVDLIMLIIFFLD 258
Query 302 GESNWLEGVMLMAAYLIISVVFWFD 326
W E ++L+ AY V F+
Sbjct 259 NVIMWWESLLLLTAYFCYVVFMKFN 283
Score = 30.0 bits (66), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 20 LGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLV 79
L TE+L + + ++ G A G+A E+ S+ + I V GT++GS + N+L V
Sbjct 161 LTVITEKLGI-SDDVAGATFMAAGGSAPELFTSLIGVFIAHSNVGIGTIVGSAVFNILFV 219
Query 80 LGM 82
+GM
Sbjct 220 IGM 222
> CE28020
Length=596
Score = 29.6 bits (65), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 32/62 (51%), Gaps = 1/62 (1%)
Query 20 LGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLV 79
L TE+LSL + ++ G A G+A E SV + I V GT++GS N+L V
Sbjct 128 LDVLTEKLSL-SDDVAGATFMAAGGSAPEFFTSVIGVFIAQNNVGIGTIVGSATFNILCV 186
Query 80 LG 81
L
Sbjct 187 LA 188
Lambda K H
0.325 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8129399586
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40